Difference between revisions of "Part:BBa K1694005"
(→Improvement of renaturing anti-Her2 scFv inclusion bodies) |
(→Improvement of renaturing anti-Her2 scFv inclusion bodies) |
||
| Line 211: | Line 211: | ||
According to our research, a team from Iran completed a series of experiments to investigate how different types, concentrations, pHs, and additives of denaturing agents affect the Inclusion bodies (IBs) solubility of HER2 scFvs. The data obtained suggested that urea 6 M solubilizes more IBs compared with other solubilizing agents. The effect of pH on the yield of IBs solubilizing was also checked out and the optimum pH was 11. Moreover, the addition of BME to the solubilizing buffer was proved to improve IBs solubilization . In all, most IBs were dissolved with urea 6 M at pH 11 containing 4 mM BME. Another set of experiments suggested that only tricin, arginine, and imidazole had positive effects while other buffer additives showed negative effects. The optimum concentrations of three buffer additives for refolding of anti-HER2 scFv were tricine, 23 mM; Arginin, 0.55 mM; And imidazole, 14.3 mM. | According to our research, a team from Iran completed a series of experiments to investigate how different types, concentrations, pHs, and additives of denaturing agents affect the Inclusion bodies (IBs) solubility of HER2 scFvs. The data obtained suggested that urea 6 M solubilizes more IBs compared with other solubilizing agents. The effect of pH on the yield of IBs solubilizing was also checked out and the optimum pH was 11. Moreover, the addition of BME to the solubilizing buffer was proved to improve IBs solubilization . In all, most IBs were dissolved with urea 6 M at pH 11 containing 4 mM BME. Another set of experiments suggested that only tricin, arginine, and imidazole had positive effects while other buffer additives showed negative effects. The optimum concentrations of three buffer additives for refolding of anti-HER2 scFv were tricine, 23 mM; Arginin, 0.55 mM; And imidazole, 14.3 mM. | ||
| + | <b>Reference:</b> | ||
| + | <ol> | ||
| + | <li>Akbari, V., Salehinia, J., Sadeghi, H. M., & Abedi, D. (2018). Improvement of solubility and refolding of an anti-human epidermal growth factor receptor 2 single-chain antibody fragment inclusion bodies. Research in Pharmaceutical Sciences, 13(6), 566. https://doi.org/10.4103/1735-5362.245968</li> | ||
| + | </ol> | ||
==Sequence and Features== | ==Sequence and Features== | ||
Revision as of 00:47, 22 October 2021
Single-chain variable fragment (Anti-HER2)
Introduction:
ScFv (Single-Chain Variable Fragment)
ScFv (single-chain variable fragment) is a fusion protein containing light (VL) and heavy (VH) variable domains connected by a short peptide linker (Fig. 1) The peptide linker (GGSSRSSSSGGGGSGGGG) is rich in glycine and serine that makes linker flexible.
Features of scFv:
1. Specific:Although remove of the constant regions , scFv still maintains the specificity of the original immunoglobulin.
2. Efficient:ScFv is smaller than the entire antibody, so that the loading of producting protein to E.coli is lower.
HER2
Human epidermal growth factor 2, abbreviated as HER2 or EtbB2, belongs to the ErbB receptor tyrosine kinase family, which is composed of four plasma membrane-bound receptor tyrosine kinases including the other three receptors, epidermal growth factor receptor(EGFR), erbB-3 (neuregulin-binding), and erbB-4.
1.Herceptin inhibition
By binding to the HER2 receptors on the surface of breast cancer cells, Herceptin prevents them from transforming the cell growth signals and slows or ceases the growth of the breast cancer cells. Moreover, Herceptin flags the HER2 receptors and strikes these receptors by the assistance of the immune system.
2.HER2 activation
HER2 is one of the members in the human epidermal growth factor receptor family. HER2 can heterodimerize with any other three receptors in the ErbB family to form the dimerization, which consequently results in the autophosphorylation and initials the signaling pathway. When the amplification or overexpression of the HER2 gene occurs, it may cause the development and progression of certain types of the breast cancer.
Reference:
1.http://www.biooncology.com/biological-pathways/her-signaling
2.http://www.cellsignal.com/contents/science-pathway-research-tyrosine-kinase/erbb-her-signaling-pathway/pathways-erbb
3.[ ]Mark M. Moasser , The oncogene HER2; Its signaling and transforming functions and its role in human cancer pathogenesis, Oncogene. Author manuscript; available in PMC 2011 January 14.
ErbB receptor family
Which are typical cell membrane receptor tyrosine kinases that are activated following ligand binding and receptor dimerization. Especially HER2 lacks a ligand, and its structure resembles a ligand-activated state and favors dimerization. The formation of dimers leads to activation of the intrinsic tyrosine kinase domain and subsequent phosphorylation on specific tyrosine residues, which serve as docking sites for a variety of molecules. Recruitment of these molecules leads to the activation of different downstream signaling cascades, including the MAPK proliferation pathway and/or the PI3K/Akt pro-survival pathway. Inappropriate signaling may occur as a result of receptor overexpression or dysregulation of receptor activation, which may lead to Increased/uncontrolled cell proliferation, decreased apoptosis (programmed cell death), enhanced cancer cell motility, and angiogenesis
The oncogene amplify or overexpress play an important role in development and progression of aggressive types of breast cancer. HER2 amplifications are seen in breast, ovarian, bladder, non-small-cell lung carcinoma, as well as several other tumor types. HER2 proteins have been shown to form clusters in cell membranes that may play a role in tumorigenesis.
Improvement of renaturing anti-Her2 scFv inclusion bodies
Group: iGEM2021_Greatbay_SCIE
Author: iGEM2021_Greatbay_SCIE
link: Greatbay_SCIE
Single chain variable fragment antibodies (scFvs) have attracted much attention due to their small size, faster bio-distribution, and better penetration into the target tissues, and ease of expression in Escherichia coli. It gave a new opportunity for modern breast cancer treatment.
As scFvs are small and non-glycosylated proteins, they can be easily overexpressed in eukaryotic hosts such as Escherichia coli (E. coli). However, highly expressed scFvs usually accumulate as unfolded protein aggregates, which are called inclusion bodies (IB). As a result, dissolving and refolding of protein from IBs become a challenging task.
According to our research, a team from Iran completed a series of experiments to investigate how different types, concentrations, pHs, and additives of denaturing agents affect the Inclusion bodies (IBs) solubility of HER2 scFvs. The data obtained suggested that urea 6 M solubilizes more IBs compared with other solubilizing agents. The effect of pH on the yield of IBs solubilizing was also checked out and the optimum pH was 11. Moreover, the addition of BME to the solubilizing buffer was proved to improve IBs solubilization . In all, most IBs were dissolved with urea 6 M at pH 11 containing 4 mM BME. Another set of experiments suggested that only tricin, arginine, and imidazole had positive effects while other buffer additives showed negative effects. The optimum concentrations of three buffer additives for refolding of anti-HER2 scFv were tricine, 23 mM; Arginin, 0.55 mM; And imidazole, 14.3 mM.
Display scFv on the cell surface of E.coli
To display the antibody outside the E.coli, we used Lipoprotein-Outer membrane protein A (Lpp-OmpA)
BBa_K1694002. According to the paper reference [1], We chose the first 9 amino acids of Lpp, and the 46~159 amino acids of OmpA.
In order to change the scFv parts easily, we added a NcoI restriction site between OmpA and scFv so that we can change various scFv DNA sequence using the NcoI restriction enzyme.
The Fig.2 showed how we combine Lpp-OmpA-N and scFv together, first we use restriction enzyme NcoI to digest the upstream and downstream parts. After ligating two digested products, there is no mixed site between Lpp-OmpA and scFv.
See this composite part:BBa_K1694015
Reference:
[1]Improving tumor targeting and therapeutic potential of Salmonella VNP20009 by displaying cell surface CEA-specific antibodies, Michal Bereta, Andrew Hayhurst, Mariusz Gajda, Paulina Chorobik, Marta Targosz, Janusz Marcinkiewicz, and Howard L. Kaufman (2007)
Experiment:
After receiving the DNA sequences from the gene synthesis company, we recombined each scFv gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each of the scFvs. The DNA sequence length of the scFvs are around 600~800 bp. In this PCR experiment, the scFv products size should be near at 850~1050 bp. The Fig.4 showed the correct size of the scFv, and proved that we successful ligated the scFv sequence onto an ideal backbone.
Application of the part:
1. Co-transform (Two plasmids)
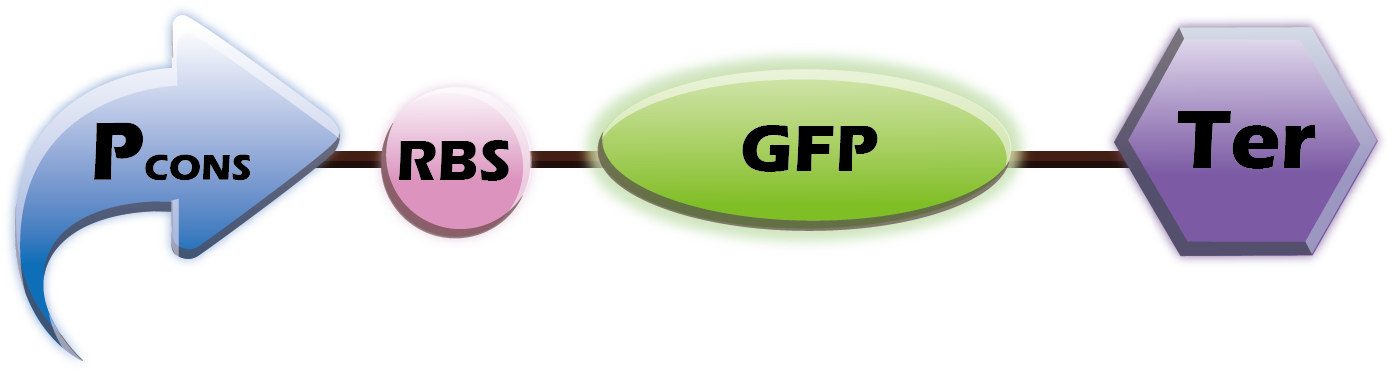
This year we want to provide a customized platform. We provide two libraries of Pcons+RBS+OmpA-scFv and Pcons+RBS+Fluorescence+Ter into E.coli. Therefore, our customers can choose any scfv and any fluorescent protein. Our team will co-transform this two plasmids, which helps us tailor our products to the wishes of our customers.
(1) Parts:
(2) Cell staining experiment:
After cloning the part of anti-HER2, we were able to co-transform anti-HER2 with different fluorescence protein into our E.coli.
The next step was to prove that our co-transformed product have successfully displayed scFv of anti-HER2 and expressed fluorescent protein.
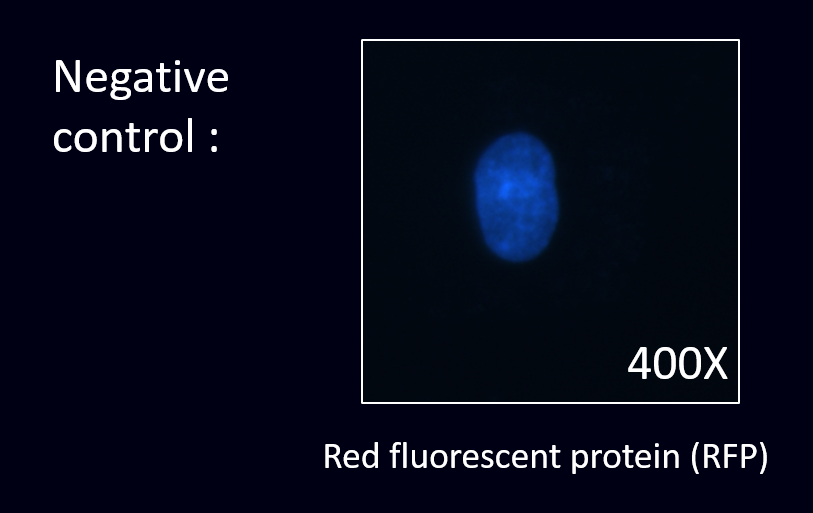
To prove this, we conducted the cell staining experiment by using the co-transformed E.coli to detect the HER2 in the cancer cell line.
(3) Staining results:
2. Transformation of single plasmid
To prove that our scFv can actually bind on to the antigen on cancer cells, we connected each scFv with a different fluorescent protein and the blue chromoprotein amilCP. Therefore we could use fluorescence microscope to clearly observe if the E.coli has produced scFv proteins. Currently, we built three different scFv connected with their respectively fluorescence protein. When applied on cell staining, we can identify the antigen distribution on cancer cells by observing the fluorescence. Furthermore, if we use the three scFv simultaneously, we can also detect multiple markers.
(1) Parts:
(2) Cell staining experiment:
After creating the part of scFv and transforming them into our E.coli, we were going to prove that our detectors have successfully displayed scFv of anti-HER2. To prove this, we have decided to undergo the cell staining experiment by using our E.coli to detect the HER2 in the SKOV-3 cancer cell lines. SKOV-3 is a kind of epithelial cell that expressed markers such as HER2.
(3) Staining results:
Modeling:
In the modeling part, we discover optimum protein expression time by using the genetic algorithm (GA) in Matlab.
We want to characterize the actual kinetics of this Hill-function based model that accurately reflects protein expression time.
By using the differential function which was derived from these optimum parameters which were calculated by GA can help us to simulate the optimum protein expression.
When we have the simulated protein expression rate, the graph of protein expression versus time can be drawn.Thus, we can find the optimum protein expression time. However, the simulated protein expression curve is slower than the experimental curve by one hour. Therefore, to find the most exact optimum protein expression time, we infer that subtracting one hour of the optimum protein expression time would be correct.
Co-transform



Transformation of single plasmid

Improvement of renaturing anti-Her2 scFv inclusion bodies
Group: iGEM2021_Greatbay_SCIE
Author: iGEM2021_Greatbay_SCIE
link: Greatbay_SCIE
Single chain variable fragment antibodies (scFvs) have attracted much attention due to their small size, faster bio-distribution, and better penetration into the target tissues, and ease of expression in Escherichia coli. It gave a new opportunity for modern breast cancer treatment.
As scFvs are small and non-glycosylated proteins, they can be easily overexpressed in eukaryotic hosts such as Escherichia coli (E. coli). However, highly expressed scFvs usually accumulate as unfolded protein aggregates, which are called inclusion bodies (IB). As a result, dissolving and refolding of protein from IBs become a challenging task.

According to our research, a team from Iran completed a series of experiments to investigate how different types, concentrations, pHs, and additives of denaturing agents affect the Inclusion bodies (IBs) solubility of HER2 scFvs. The data obtained suggested that urea 6 M solubilizes more IBs compared with other solubilizing agents. The effect of pH on the yield of IBs solubilizing was also checked out and the optimum pH was 11. Moreover, the addition of BME to the solubilizing buffer was proved to improve IBs solubilization . In all, most IBs were dissolved with urea 6 M at pH 11 containing 4 mM BME. Another set of experiments suggested that only tricin, arginine, and imidazole had positive effects while other buffer additives showed negative effects. The optimum concentrations of three buffer additives for refolding of anti-HER2 scFv were tricine, 23 mM; Arginin, 0.55 mM; And imidazole, 14.3 mM.
Reference:
- Akbari, V., Salehinia, J., Sadeghi, H. M., & Abedi, D. (2018). Improvement of solubility and refolding of an anti-human epidermal growth factor receptor 2 single-chain antibody fragment inclusion bodies. Research in Pharmaceutical Sciences, 13(6), 566. https://doi.org/10.4103/1735-5362.245968
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 234
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]