Difference between revisions of "Part:BBa K3716012"
| Line 17: | Line 17: | ||
# Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 4ml LB medium with kanamycin. Add 1mM iPTG to all experimental groups. Incubate at 37℃ in a shaker overnight. | # Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 4ml LB medium with kanamycin. Add 1mM iPTG to all experimental groups. Incubate at 37℃ in a shaker overnight. | ||
# Add 100 µl bacteria culture medium into a sterile 96-well plate. Measure fluorescence at 488nm excitation and 502nm absorption, and then also OD600 for normalization, with a microplate reader. | # Add 100 µl bacteria culture medium into a sterile 96-well plate. Measure fluorescence at 488nm excitation and 502nm absorption, and then also OD600 for normalization, with a microplate reader. | ||
| − | |||
# SDS-PAGE. The cells are lysed by 150 μL 4% SDS for 5 min at room temperature, then 10 min at 95℃. Then 6x loading buffer is added. The SDS-PAGE was carried with a 12% precast polyacrylamide gel. Coomassie bright blue stain is used to observe the bands. | # SDS-PAGE. The cells are lysed by 150 μL 4% SDS for 5 min at room temperature, then 10 min at 95℃. Then 6x loading buffer is added. The SDS-PAGE was carried with a 12% precast polyacrylamide gel. Coomassie bright blue stain is used to observe the bands. | ||
# | # | ||
| Line 29: | Line 28: | ||
| − | ''' | + | '''2. Successful Expression of the Protein with SDS-PAGE''' |
| − | + | ||
| − | + | [[File:T--iBowu-China--2021lacI-4.png|thumb|center|600px|'''Figure.3. Expression of β-glucuronidase withIptg induction. The red arrow marked the expected size band around 67 kDa where the enzyme is expected to be located. Control groups where there is no Iptg induction for the same plasmid and where the sequence was completely removed (labeled Pet) showed no intrinsic protein exists in the expected bands.''' ]] | |
| + | |||
| + | To express the enzyme β-glucuronidase, the plasmid was clone in DH5a and then extracted to transform BL21(DE3). Iptg was added at 1mM at OD600=0. The medium was incubated at 37 degree Celsius with shaking overnight, and then taken to SDS-PAGE experiment to check for the expression of the enzyme. This enzyme, which we named T7-bG-pet, was expected to have a size about 67 kDa, and the SDS-PAGE result showed the successful expression of the protein at the expected size band (marked by a red arrow on the figure). We included a control group of vacant-pet28a, of which the only difference is there the β-glucuronidase sequence is removed. Control groups where there is no Iptg induction for the same plasmid and control groups of vacant-pet28a (labeled Pet) showed no intrinsic protein exists in the expected bands. | ||
Revision as of 05:11, 18 October 2021
6xHis-Beta-glucuronidase (E.coli, gusA)
6xHis-Beta-glucuronidase (E.coli, gusA)
Design
Introduction
This year team iBowu-China used this part to express the enzyme called β-glucuronidase in E. coli strain BL21(DE3) and similar variants of the strain. The designed purpose of this enzyme is to convert glycyrrhizic acid into glycyrrhetinic acid, which is the effective active ingredient of licorice.
Protocol
- Transform the plasmids into E. coli BL21(DE3)
- Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 4ml LB medium with kanamycin. Add 1mM iPTG to all experimental groups. Incubate at 37℃ in a shaker overnight.
- Add 100 µl bacteria culture medium into a sterile 96-well plate. Measure fluorescence at 488nm excitation and 502nm absorption, and then also OD600 for normalization, with a microplate reader.
- SDS-PAGE. The cells are lysed by 150 μL 4% SDS for 5 min at room temperature, then 10 min at 95℃. Then 6x loading buffer is added. The SDS-PAGE was carried with a 12% precast polyacrylamide gel. Coomassie bright blue stain is used to observe the bands.
Results
1. Confirmation of the Gene Circuit with sfGFP
First we needed to experimentally confirm that our gene circuit would work as we had expected. We used sfGFP (Part:K515105) gene to substitute the β-glucuronidase gene in the above plasmid construction. The reconstructed plasmid was then cloned in DH5a and then extracted to be transformed into BL21(DE3). With the induction Iptg added, we can clearly measure strong signal of green fluorescence light. The intensity is about 100 fold stronger than the background, which is obtained by measuring the same bacteria culture but without Iptg induction. This result confirm that with Iptg induction, our gene circuit works normally and can be used for expression of bG enzyme.
2. Successful Expression of the Protein with SDS-PAGE
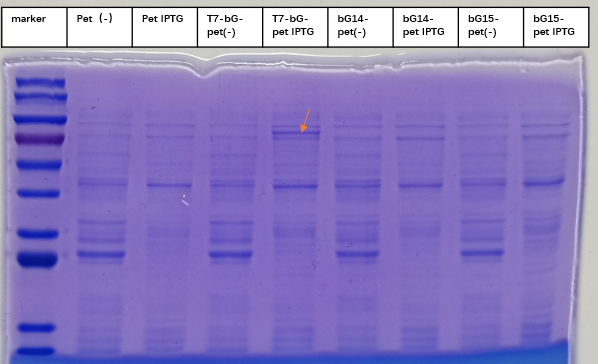
To express the enzyme β-glucuronidase, the plasmid was clone in DH5a and then extracted to transform BL21(DE3). Iptg was added at 1mM at OD600=0. The medium was incubated at 37 degree Celsius with shaking overnight, and then taken to SDS-PAGE experiment to check for the expression of the enzyme. This enzyme, which we named T7-bG-pet, was expected to have a size about 67 kDa, and the SDS-PAGE result showed the successful expression of the protein at the expected size band (marked by a red arrow on the figure). We included a control group of vacant-pet28a, of which the only difference is there the β-glucuronidase sequence is removed. Control groups where there is no Iptg induction for the same plasmid and control groups of vacant-pet28a (labeled Pet) showed no intrinsic protein exists in the expected bands.
1. Confirmation of the Gene Circuit with sfGFP
图:酶活图
大肠杆菌的bG酶具有较好的酶活,而米曲霉和古生菌的bG酶有文献报道具有较好的酶活,但我们发现其在大肠杆菌中活性较低。
Summary
LacI can effectively work together with LacO to control gene expression in E. coli BL21(DE3). Our results confirmed 0.5 to 2mM Iptg can be effective and sufficient to release the suppression of gene expression exerted by LacI protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]


