Difference between revisions of "Part:BBa K302033"
| Line 48: | Line 48: | ||
===Characterized by XJTU-China 2020=== | ===Characterized by XJTU-China 2020=== | ||
| + | ===Characterization=== | ||
| + | This year, our team designed a suicide switch mediated by arabinose operon for biosafety. We use MazF to kill the bacteria. We have supplemented the relevant information of this element in the iGEM Parts and added test and characterization to it. | ||
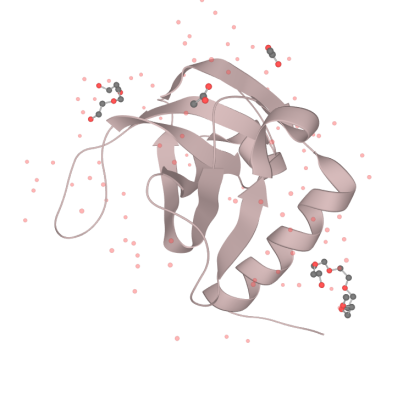
| + | <div>[[File:T--XJTU-China--Contributation01.png |700px|thumb|center|<b>Figure 1:</b> the structure of MazF.]]</div> | ||
| + | Our MazF comes from the genome of Bacillus Subtilis. By regulating the expression of MazF, we inhibit the life of bacteria when needed to prevent the possible strains and genetic leakage. | ||
| + | MazF is a kind of mRNA interferas. Specifically, it specifically cleaves five-base U^ACAU sequence (^indicating the cleavage site) on RNA[1]. MazF achieves its catalytic function by forming a special MazF-RNA complex. One molecule of 9-mer RNA is bound to one MazF dimer, and two subunits of MazF form the dimer related by local 2-fold symmetry.The RNA in an extended alignment is bound along the RNA binding interface between subunits of the MazF dimer, covering part of this dimeric interface, and MazF interacts extensively with the pentad target sequence present in the RNA chain by interactions with bases[2]. Therefore, the backbone phosphate moieties project outward and away from the protein surface. In this way, scissile phosphate will be attacked by other side chain groups, which causes breaking of P-O bond. | ||
| + | What’s more, it is an important part of the Bacillus Subtilis toxin-antitoxin system. Which is essential for the programmed cell death of this developmental bacterium. In normally growing cells, MazF forms a stable complex with its cognate antitoxin, MazE, however, under stress conditions, unstable MazE is preferentially degraded to release free MazF in the cells, which then cleaves cellular mRNAs to inhibit protein synthesis, leading to growth arrest. The protein-protein interaction interface between MazE and MazF (2,843 Å2) is larger than the MazF-RNA interaction interface (2,153 Å2), suggesting that MazF is likely to have a higher affinity for MazE over its RNA substrate. In the B.S MazE-MazF complex structure, MazE binds to MazF along the dimer interface, and it even occupies part of the putative modeled RNA binding site on the second subunit of MazF[3]. Thus, in the presence of MazE, MazF cannot bind to or cleave substrate RNA. | ||
| + | |||
| + | Biochemical properties of MazF | ||
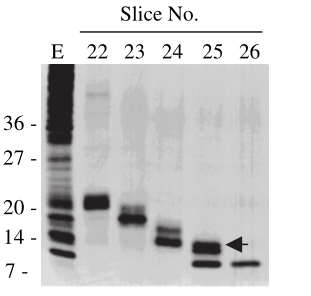
| + | <div>The size of MazF is about14 kDa. The enzyme activity showed very little pH dependence between pH 5.0 and 9.0 in either 10 mM T ris-maleate or 10 mM citrate-phosphate buffer. It was also active over a wide range of temperatures from 4℃ to 50℃,with an optimum of around 28℃. The enzyme is inhibited by high salt concentrations and optimal enzyme activity was seen in the complete absence of any | ||
| + | divalent cation[4]. | ||
| + | <div>[[File:T--XJTU-China--Contributation02.png |700px|thumb|center|<b>Figure 2:</b> Material eluted, acetone precipitated and reapplied to an SDS-20% polyacrylamide gel and then silver stained. Slice numbers are the same as in A. E is whole cell extract used as starting material for protein elution. Arrow shows the estimated position of MazF migration. | ||
| + | |||
| + | Ref: Pellegrini O, Mathy N, Gogos A, Shapiro L, Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol. 2005 Jun;56(5):1139-48. doi: 10.1111/j.1365-2958.2005.04606.x. PMID: 15882409.]]</div> | ||
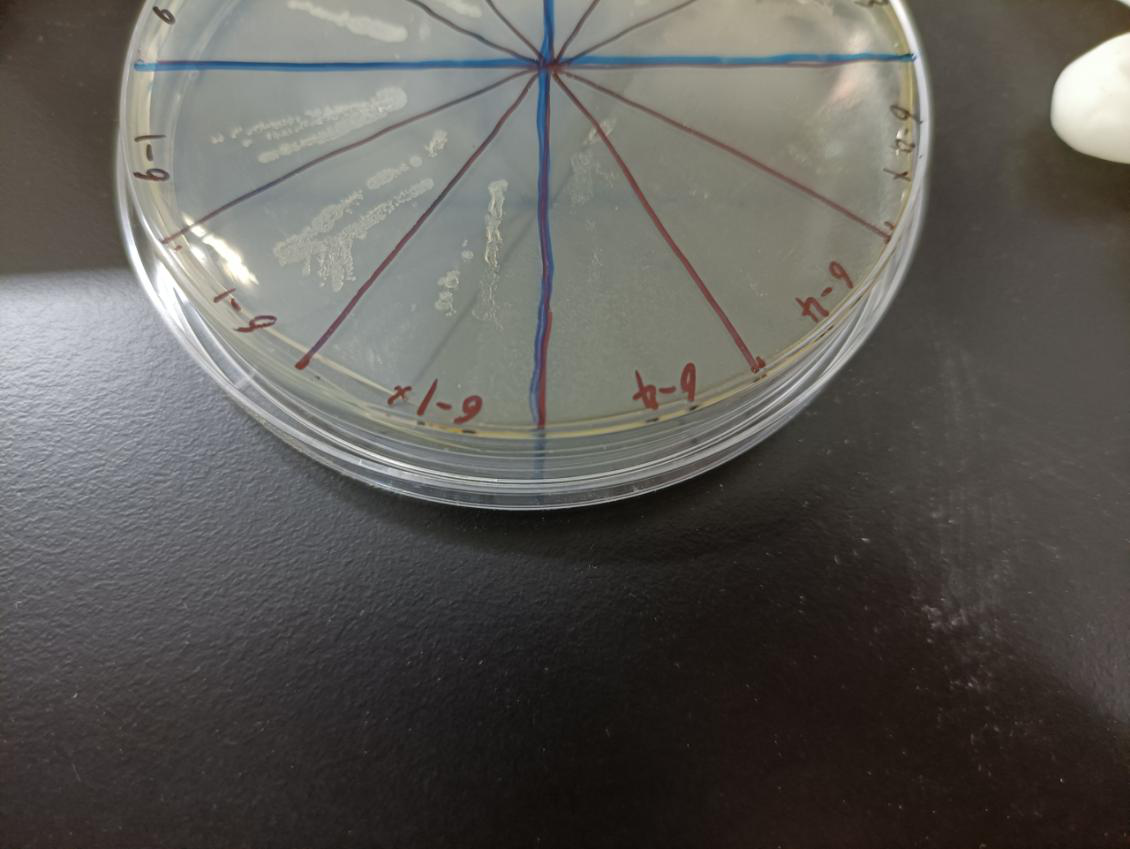
In the figure, the bacteria in this plate contained plasmids of pBAD-MazF. Three of 6-1 | In the figure, the bacteria in this plate contained plasmids of pBAD-MazF. Three of 6-1 | ||
groups were the control group without arabinose induction, and three of 6-4 groups were | groups were the control group without arabinose induction, and three of 6-4 groups were | ||
| Line 56: | Line 69: | ||
Result: The bacteria without arabinose induction grow normally, while with arabinose | Result: The bacteria without arabinose induction grow normally, while with arabinose | ||
induction, the bacteria suicide. | induction, the bacteria suicide. | ||
| − | <div>[[File:T--XJTU-China--Implementation03.png |700px|thumb|center|<b>Figure | + | <div>[[File:T--XJTU-China--Implementation03.png |700px|thumb|center|<b>Figure 3:</b> Experimental medium. 6-1 is blank control ,6-4 is experimental group.]]</div> |
| + | |||
| + | ==Reference== | ||
| + | [1]Park, Jung-Ho, Yamaguchi, Yoshihiro and Inouye, Masayori(2011), Bacillus subtilis MazF‐bs (EndoA) is a UACAU‐specific mRNA interferase, FEBS Letters, 585, doi: 10.1016/j.febslet.2011.07.008 | ||
| + | [2]Dhirendra K. Simanshu, Yoshihiro Yamaguchi, Jung-Ho Park, et al. Structural Basis of mRNA Recognition and Cleavage by Toxin MazF and Its Regulation by Antitoxin MazE in Bacillus subtilis. 2013, 52(3):447-458 | ||
| + | [3]Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 2005; 56:845–857. [PubMed: 15853875] | ||
| + | [4] Pellegrini O, Mathy N, Gogos A, Shapiro L, Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol. 2005 Jun;56(5):1139-48. doi: 10.1111/j.1365-2958.2005.04606.x. PMID: 15882409. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 00:43, 28 October 2020
mazF
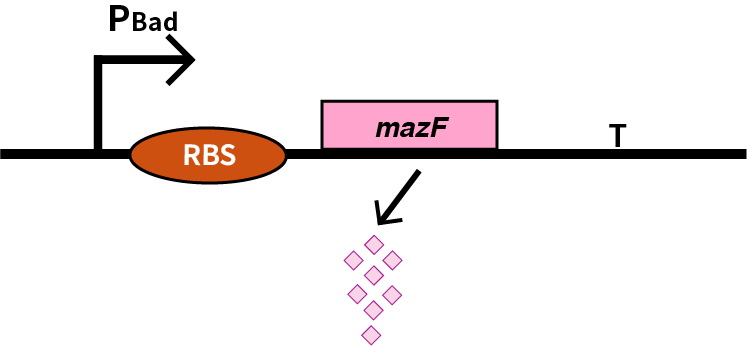
Encodes a stable non-specific ribonuclease in Bacillus Subtilis. It is used in conjungtion with mazE in Bacillus Subtilis to provide a toxin-antitoxin kill switch in various stressful conditions. When expression of both genes is turned off (as both are under contol of same promoter) mazE will be degraded faster than mazF. There is then no inhibiton of mazF, killing the cell.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterized by BNU-China 2019
We characterize mazF (BBa_K302033) by an induced suicide system, in which the downstream mazF (BBa_K302033) gene encoding toxin MazF is put under the control of L-arabinose induced promoter pBAD (BBa_K206000). MazF is an endoribonuclease that cleaves RNAs at ACA sites and causes the death of microbe as a reporter [1]. As a result, we can characterize mazF in a cell density-dependent manner in Escherichia coli K-12.
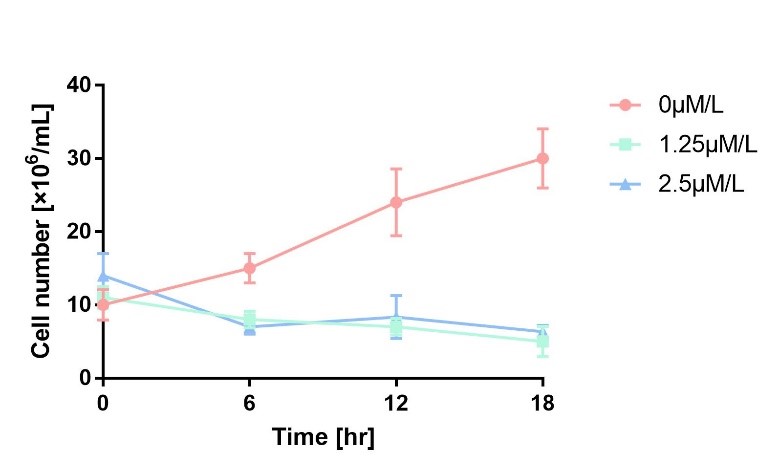
In order to characterize the toxicity of protein encoded by mazF, we take engineered microbe without induction as control group. As is shown in Fig.1, the cell number of experimental groups show a significant decrease upon induction, which indicates the protein encoded by mazF is lethal to E. coli.
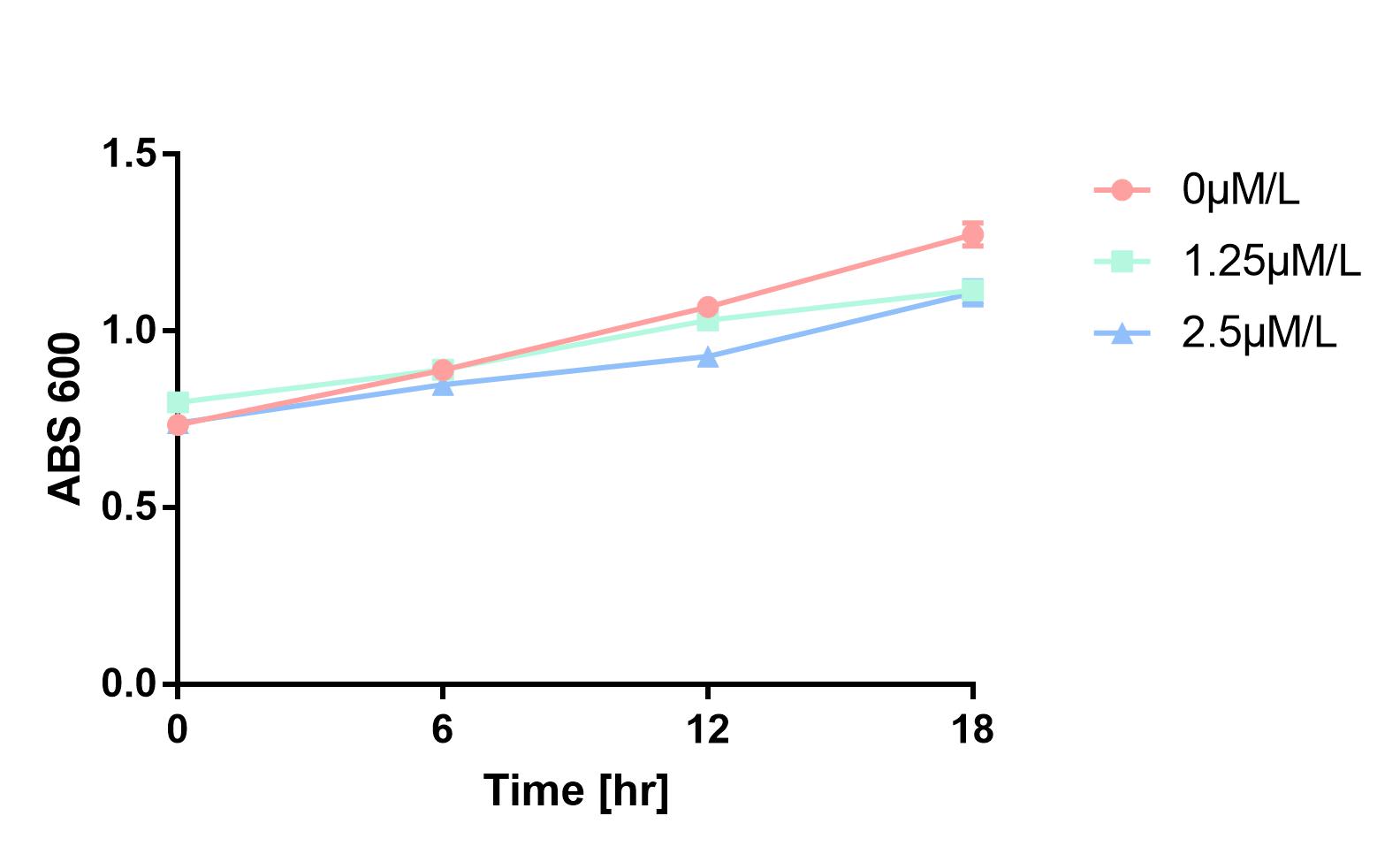
Furthermore, in order to prove that MazF kills the cell without lysing it, we measure OD600 of each sample to give an overall number of intact bacteria, dead and alive. As is shown in Fig.2, there is little difference between control and experimental groups, although it is validated numbers of alive cells differ. Hence, we reach a conclusion that protein encoded by mazF mediates suicide of cells without causing lysis of bacterial cells.
Experimental approach
1. Transform the plasmids into E. coli DH5α competent cells.
2. The engineered bacteria are cultured in 200mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm;
3. Equally divide the culture into 90 centrifuge tubes, which is 1mL respectively. Centrifuge them at 4000rpm for 5 minutes. Discard the liquid.
4. Resuspend 30 tubes of collected bacteria with LB-ampicillin (50 ng/µl) containing 1.25μM/L and 2.5μM/L L-arabinose respectively as experimental groups. Resuspend 30 tubes of bacteria with pure LB-ampicillin (50 ng/µl) medium.
5. Collect 3 tubes of all groups every 6 hours, dilute all of the samples to 10^7 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately. At the same time, refresh the medium to maintain the concentration of L-arabinose.
6. Count the number of colonies in 5 cm^2 per plate after cultured for 24 hours at 37℃
7. Three repicas are tested in each group.
Reference
[1] Diana Széliová, Ján Krahulec, Martin Šafránek, et al. Modulation of heterologous expression from PBAD promoter in Escherichia coli production strains[J]. Journal of Biotechnology, 2016, 236:1-9.
Characterized by XJTU-China 2020
Characterization
This year, our team designed a suicide switch mediated by arabinose operon for biosafety. We use MazF to kill the bacteria. We have supplemented the relevant information of this element in the iGEM Parts and added test and characterization to it.
Our MazF comes from the genome of Bacillus Subtilis. By regulating the expression of MazF, we inhibit the life of bacteria when needed to prevent the possible strains and genetic leakage. MazF is a kind of mRNA interferas. Specifically, it specifically cleaves five-base U^ACAU sequence (^indicating the cleavage site) on RNA[1]. MazF achieves its catalytic function by forming a special MazF-RNA complex. One molecule of 9-mer RNA is bound to one MazF dimer, and two subunits of MazF form the dimer related by local 2-fold symmetry.The RNA in an extended alignment is bound along the RNA binding interface between subunits of the MazF dimer, covering part of this dimeric interface, and MazF interacts extensively with the pentad target sequence present in the RNA chain by interactions with bases[2]. Therefore, the backbone phosphate moieties project outward and away from the protein surface. In this way, scissile phosphate will be attacked by other side chain groups, which causes breaking of P-O bond.
What’s more, it is an important part of the Bacillus Subtilis toxin-antitoxin system. Which is essential for the programmed cell death of this developmental bacterium. In normally growing cells, MazF forms a stable complex with its cognate antitoxin, MazE, however, under stress conditions, unstable MazE is preferentially degraded to release free MazF in the cells, which then cleaves cellular mRNAs to inhibit protein synthesis, leading to growth arrest. The protein-protein interaction interface between MazE and MazF (2,843 Å2) is larger than the MazF-RNA interaction interface (2,153 Å2), suggesting that MazF is likely to have a higher affinity for MazE over its RNA substrate. In the B.S MazE-MazF complex structure, MazE binds to MazF along the dimer interface, and it even occupies part of the putative modeled RNA binding site on the second subunit of MazF[3]. Thus, in the presence of MazE, MazF cannot bind to or cleave substrate RNA.
Biochemical properties of MazF
divalent cation[4].
In the figure, the bacteria in this plate contained plasmids of pBAD-MazF. Three of 6-1 groups were the control group without arabinose induction, and three of 6-4 groups were the experimental group with 1.5 M/L arabinose induction. Where, the dilution factor of the group with * is 10^4, and the dilution factor of the group without * is 10^6.
Result: The bacteria without arabinose induction grow normally, while with arabinose induction, the bacteria suicide.
Reference
[1]Park, Jung-Ho, Yamaguchi, Yoshihiro and Inouye, Masayori(2011), Bacillus subtilis MazF‐bs (EndoA) is a UACAU‐specific mRNA interferase, FEBS Letters, 585, doi: 10.1016/j.febslet.2011.07.008 [2]Dhirendra K. Simanshu, Yoshihiro Yamaguchi, Jung-Ho Park, et al. Structural Basis of mRNA Recognition and Cleavage by Toxin MazF and Its Regulation by Antitoxin MazE in Bacillus subtilis. 2013, 52(3):447-458 [3]Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 2005; 56:845–857. [PubMed: 15853875] [4] Pellegrini O, Mathy N, Gogos A, Shapiro L, Condon C. The Bacillus subtilis ydcDE operon encodes an endoribonuclease of the MazF/PemK family and its inhibitor. Mol Microbiol. 2005 Jun;56(5):1139-48. doi: 10.1111/j.1365-2958.2005.04606.x. PMID: 15882409.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]