Difference between revisions of "Part:BBa K3595004"
Liyingying (Talk | contribs) |
Liyingying (Talk | contribs) |
||
| Line 10: | Line 10: | ||
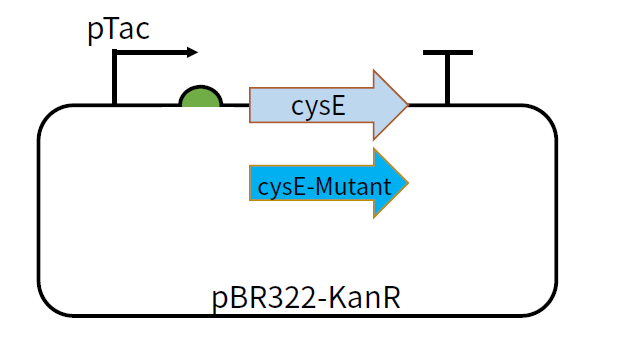
[[File:T--GZ_HFI--cysE.png|600px|thumb|center|The structure of the plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant ]] | [[File:T--GZ_HFI--cysE.png|600px|thumb|center|The structure of the plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant ]] | ||
==Experimental Setup== | ==Experimental Setup== | ||
| − | *Genetic information of cysE,cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2 was described on the page of [[Part:BBa_K3595005]],[[Part:BBa_K3595006]],[[Part:BBa_K3595007]], [[Part:BBa_K3595008]],[[Part:BBa_K3595009]], [[Part: | + | *Genetic information of cysE,cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2 was described on the page of [[Part:BBa_K3595005]],[[Part:BBa_K3595006]],[[Part:BBa_K3595007]], [[Part:BBa_K3595008]],[[Part:BBa_K3595009]], [[Part:BBa_K3595010]], [[Part:BBa_K3595011]],respectively. |
*Plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant was transfered into the <i>Nissle </i> host cell,respestively. | *Plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant was transfered into the <i>Nissle </i> host cell,respestively. | ||
*Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | *Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | ||
Revision as of 20:58, 27 October 2020
cysE-Wildtype
The wild-type cysE gene encodes L-serine O-acetyl transferase (SAT), which is a key enzyme in the pathway of hydrogen sulfide metabolism.In the pathway of converting hydrogen sulfide to L-cysteine, L-serine and Acetyl-CoA form O-acetylserine catalyzed by SAT.
Usage and Biology
This part can be used as a coding sequence after the promoter pTac and RBS B0034. The feedback inhibition-insensitive SAT can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-Mutant,among which the mutants include cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2. The constructed plasmid was transformed into Nissle host cell to test its production of cysteine.
Experimental Setup
- Genetic information of cysE,cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2 was described on the page of Part:BBa_K3595005,Part:BBa_K3595006,Part:BBa_K3595007, Part:BBa_K3595008,Part:BBa_K3595009, Part:BBa_K3595010, Part:BBa_K3595011,respectively.
- Plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant was transfered into the Nissle host cell,respestively.
- Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm.
- Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.The media contained 3 ul kanamycin and 1.5 uL 1M IPTG. At the same time, wild-type Nissle was inoculated as negative control, and M9 medium was used as blank control.
- Detecting cysteine concentration in culture medium
Results
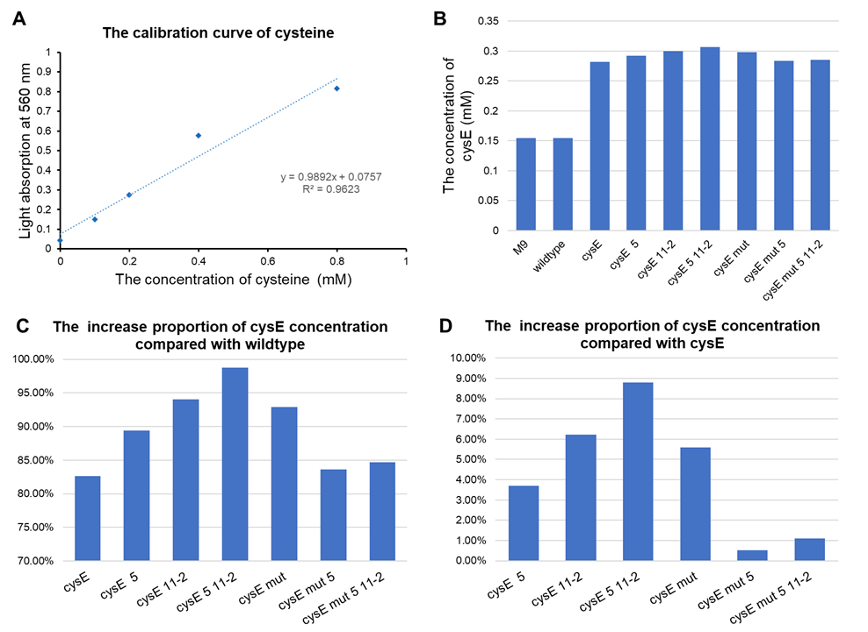
- All of our engineered bacteria have shown great improvement in the ability to produce cysteine, which represents that they can absorb H2S to a larger extent.
- EcN with plasmid pTYT-cysE-5-11-2 had the best effect, 98.72% batter than EcN wildtype and 8.79% better than overexpression of cysE (pTYT-cysE)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 684
- 1000COMPATIBLE WITH RFC[1000]
Reference
Kondoh M, et al. L-Cysteine production by metabolically engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol 2019;103(6):2609-19.