Difference between revisions of "Part:BBa K1529321:Experience"
(→Contribution: Waseda 2020) |
(→Contribution: Waseda 2020) |
||
| Line 74: | Line 74: | ||
]]<br> | ]]<br> | ||
| − | [[Image:T--Waseda--crosstalk_rhlR.png|thumb|center|600px|<b>Fig. | + | [[Image:T--Waseda--crosstalk_rhlR.png|thumb|center|600px|<b>Fig. 2. </b>Crosstalk assay in a cell-free system containing rhlR protein. |
]]<br> | ]]<br> | ||
Latest revision as of 05:40, 27 October 2020
Prhl(RR)_GFP
Contents
Materials and Methods
1. Construction
All the samples were JM2.300 strain with antibiotic resistance to ampicillin and kanamycin.
A. Ptet_RhlR (6A1) Prhl-GFP (3K3)
B. Ptet_RhlR (6A1) Prhl(RR)-GFP (3K3)
C. Ptet_RhlR (6A1) Prhl(LR)-GFP (3K3)
D. Ptet_RhlR (6A1) Prhl(RL)-GFP (3K3)
E. Ptet_RhlR (6A1) Placuv5-GFP (3K3) …positive control
F. Ptet_RhlR (6A1) promoter less-GFP(3K3) …negative control
2. Assay protocol
1.Prepare 2 overnight cultures for each sample A~F in 3 mL LB medium, containing ampicillin (50 microg / mL) and kanamycin (30 microg / mL) at 37°C for 12 h.
2.Dilute the overnight cultures to 1 / 100 in fresh LB medium (3 mL) containing ampicillin (50 microg / mL) and kanamycin (30 microg / mL) (→fresh culture).
3.Incubate the fresh cultures in 37°C until the OD590 reaches 0.3.
4.Add 30 microL of 500 microM C4HSL or DMSO as listed below:
A-5 microM: A + C4HSL
A-0 microM: A + DMSO
B-5 microM: B + C4HSL
B-0 microM: B + DMSO
C-5 microM: C + C4HSL
C-0 microM: C + DMSO
D-5 microM: D + C4HSL
D-0 microM: D + DMSO
E-5 microM: E + C4HSL
E-0 microM: E + DMSO
F-5 microM: F + C4HSL
F-0 microM: F + DMSO
5.Incubate the samples at 37°C for 4 h.
6.Start preparing the flow cytometer 1 h before the end of incubation.
7.Take 200 microL of the sample, and centrifuge at 9000x g, 1 min., 4°C.
8.Remove the supernatant by using P1000 pipette.
9.Add 1 mL of filtered PBS (phosphate-buffered saline) and suspend.
10.Dispense all of each suspension into a disposable tube through a cell strainer.
11.Measure fluorescence intensity with a flow cytometer (We used BD FACSCaliburTM Flow Cytometer of Becton, Dickenson and Company).
Results
Discussion
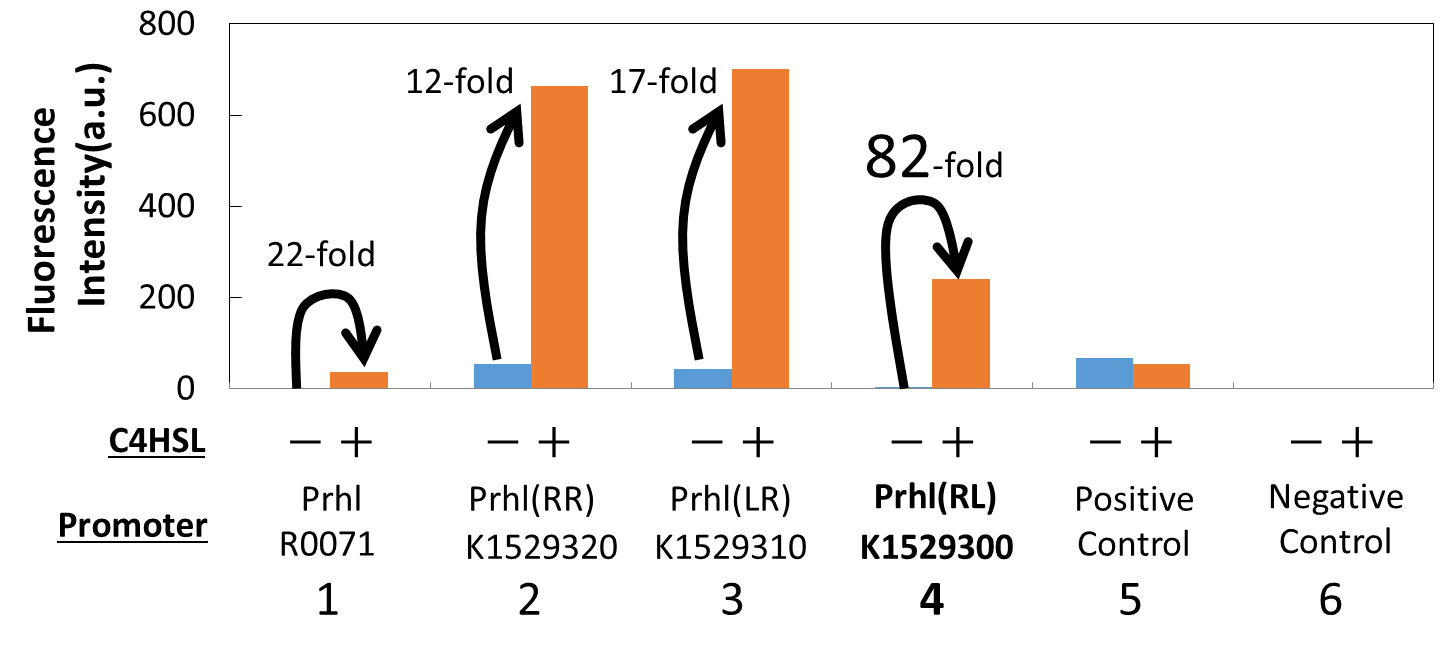
We measured the GFP expression with the four different promoters (Prhl : BBa_R0071, Prhl(RR) : BBa_K1529320, Prhl(LR) : BBa_K1529310, and Prhl(RL) : BBa_K1529300) by flow cytometer. Each promoter was tested in the presence and also in the absence of C4HSL (See Materials and Methods for detailed procedures).
Fig. 2. shows the fluorescence intensity detected by flow cytometer.
Fig. 3. is the extracted data which shows the comparison of the promoters: Prhl, Prhl(RR), and Prhl(RL).
As Fig. 3 shows, when induced by C4HSL, Prhl(RR) promoter showed higher maximum expression level and higher leak than the original Prhl promoter.
Although Prhl(RL) promoter had lower maximum expression level compared to Prhl(RR) promoter, it had the highest induced/not-induced ratio.
This means Prhl(RL) promoter has little leak.
Therefore, we can say that Prhl(RL) promoter is the best improved Prhl promoter due to the advantages of less leak and higher expression level.
For more information, see [http://2014.igem.org/Team:Tokyo_Tech/Experiment/Prhl_reporter_assay our work in Tokyo_Tech 2014 wiki].
Applications of BBa_K1529321
Contribution: Waseda 2020
For the crosstalk experiments, we constructed two kinds of cell-free translation systems containing substantial amount of R proteins (LuxR,RhlR). In those systems, we put signaling molecules (3OHSL-C4 or 3OHSL-C6) and target genes (Plux/tet-GFP or Prhl-RR-GFP), respectively. We measured the fluorescence of GFP expressed from plux/tet-GFP and prhl(RR)-GFP on a RT-qPCR machine.
Fig 1 shows the result of the crosstalk test in a cell-free system containing sufficient amount of LuxR protein. In the results, non-cognate Prhl(RR) promter was highly activated by luxR-3OC6HSL complex as well as the cognate Plux/tet promoter. Fig 2 shows the result of the crosstalk test in a cell-free system containing sufficient amount of RhlR protein. As a result, we revealed that Prhl(RR)-GFP can work in vitro.
User Reviews
UNIQ6b0d0d68f92ab1c5-partinfo-00000005-QINU UNIQ6b0d0d68f92ab1c5-partinfo-00000006-QINU