Difference between revisions of "Part:BBa K3593000"
| Line 5: | Line 5: | ||
Aptamer of alpha amatoxin found by iterative SELEX process. It forms special secondary structure and bind with amatoxin specifically, which is characterized by q PCR | Aptamer of alpha amatoxin found by iterative SELEX process. It forms special secondary structure and bind with amatoxin specifically, which is characterized by q PCR | ||
=Background= | =Background= | ||
| + | Amatoxins are chemicals present inside the genus Amanita and caused about 90% of mushroom poisoning. Being able to detect it before eating or in the field could possibly make a great decrease in people and animals who died because of poisonous mushrooms. Also being able to detect it in hospital can greatly help doctors in mushroom areas get correct information and do effective diagnosis to save the patient. | ||
| + | |||
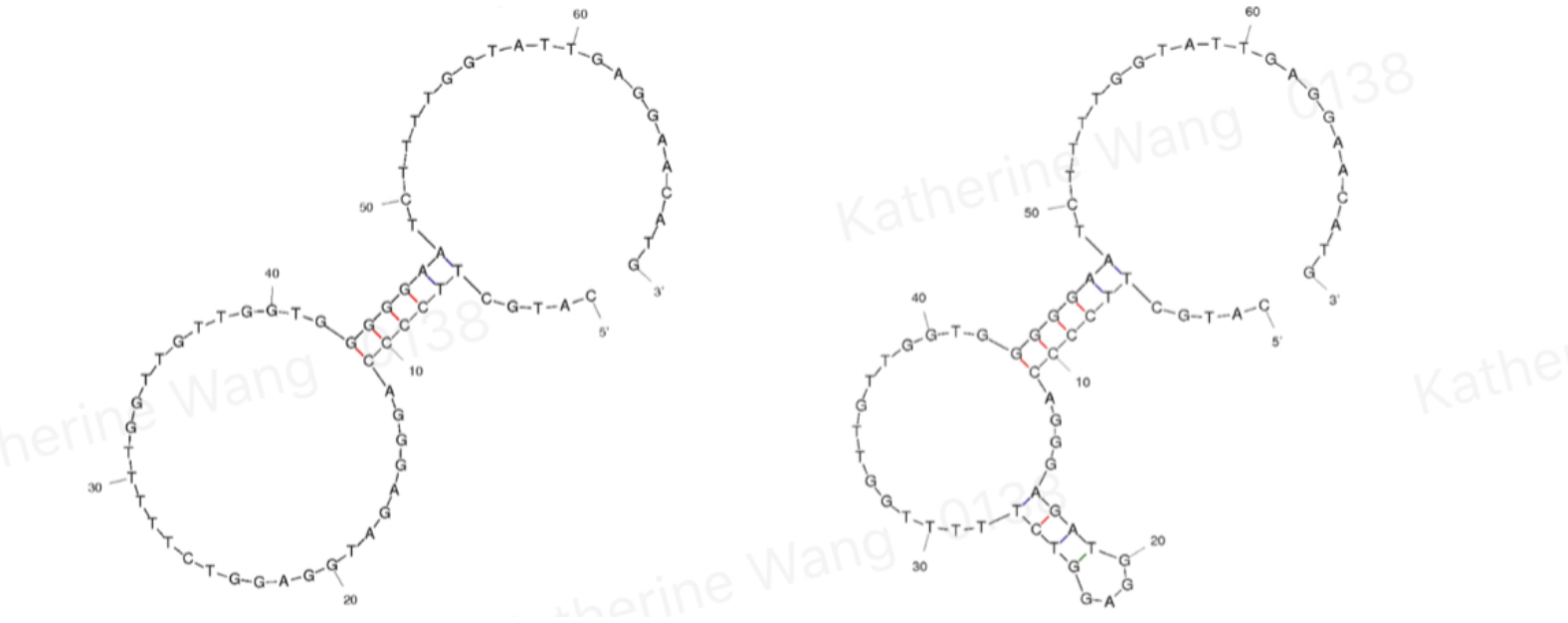
| + | Aptamers are oligonucleotides that form secondary structures, giving them the ability to bind targeted molecules, including ions or small molecules, and, in our case, amanitin. BBa_K3593000 is an aptamer of α-amanitin named Best 1. It comes from one previous literature[1]. Its binding is tested through qPCR. | ||
| + | [[File:T--GreatBay_SCIE--Fig.1 Structure of aptamer Best 1 and Best 2.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Fig.1 Structure of aptamer Best 1 and Best 2</center></b></p> | ||
=Sequence and features= | =Sequence and features= | ||
<partinfo>BBa_K3593000 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3593000 SequenceAndFeatures</partinfo> | ||
=Characterisation of this part= | =Characterisation of this part= | ||
| − | |||
==Experiment data== | ==Experiment data== | ||
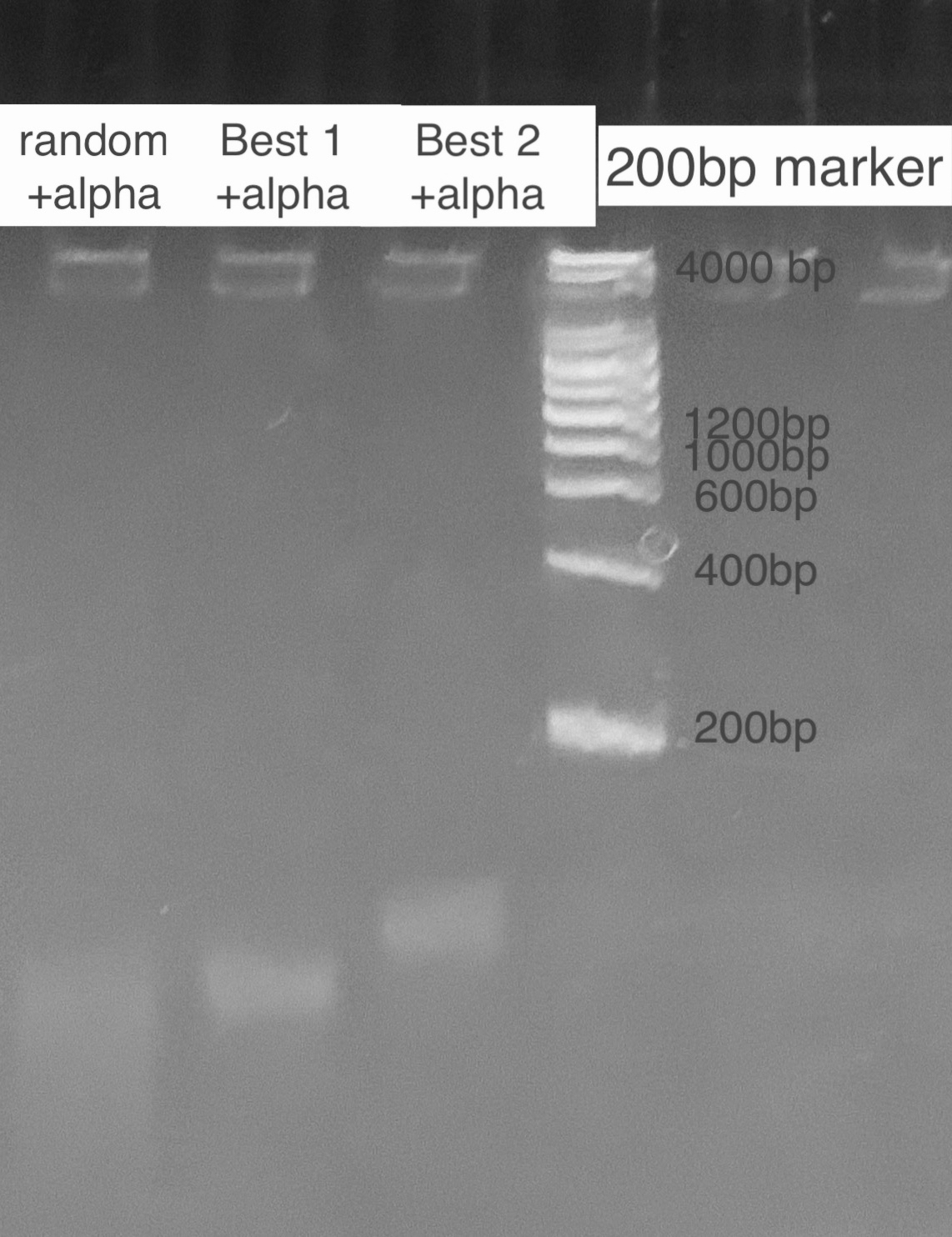
| + | According to the state of art in aptamer binding[3], we design an experiment based on the principle of Gel Shift(i.e lower rate of motion in electrophoresis when aptamer is bound to the toxin due to a conformational change). By mixing excess amatoxin determined by the calculation of dissociation constant and aptamer in certain reaction times, we expect to see 2 bands representing the bound and inbound aptamer respectively. For examination of specificity, we also mix a random ssDNA pool where we select aptamer from and compare the band with the correct aptamer. For a high resolution, we used the TBE PAGE gel for DNA, 12%. <br> | ||
| + | [[File:T--GreatBay_SCIE--Fig.2 Gel Shift of Best 1&Best 2.jpg|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Fig.2 Gel Shift of Best 1&Best 2</center></b></p> | ||
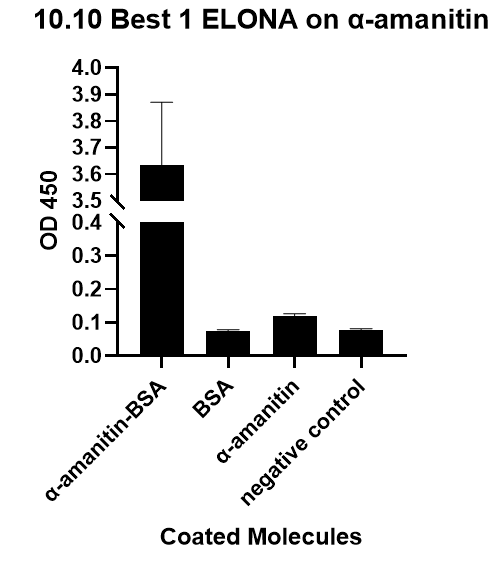
| + | We also verify its binding affinity via a method called ELONA(enzyme-linked oligonucleotide assay), a method | ||
| + | from another literature[2], with a similar principle to ELISA.<br> | ||
| + | [[File:T--GreatBay_SCIE--Fig.3 The ELONA on Best 1 aptamer.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Fig.3 The ELONA on Best 1 aptamer.</center></b></p> | ||
=References= | =References= | ||
| − | + | 1.Muszyńska K, Ostrowska D, Bartnicki F, et al. Selection and analysis of a DNA aptamer binding α-amanitin from Amanita phalloides. Acta Biochim Pol. 2017;64(3):401-406. | |
| + | 2.Han Q, Xia X, Jing L, et al. Selection and characterization of DNA aptamer specially targeting α-amanitin in wild mushrooms. SDRP J Food Sci Technol. 2018;3(6):497-508. | ||
| + | 3.Muszyńska K, Ostrowska D, Bartnicki F, et al. Selection and analysis of a DNA aptamer binding α-amanitin from Amanita phalloides. Acta Biochim Pol. 2017;64(3):401-406. doi:10.18388/abp.2017_1615 | ||
<!-- Add more about the biology of this part here> | <!-- Add more about the biology of this part here> | ||
Revision as of 02:47, 27 October 2020
ssDNA, aptamer for α-amanitin(Best 1)
Aptamer of alpha amatoxin found by iterative SELEX process. It forms special secondary structure and bind with amatoxin specifically, which is characterized by q PCR
Background
Amatoxins are chemicals present inside the genus Amanita and caused about 90% of mushroom poisoning. Being able to detect it before eating or in the field could possibly make a great decrease in people and animals who died because of poisonous mushrooms. Also being able to detect it in hospital can greatly help doctors in mushroom areas get correct information and do effective diagnosis to save the patient.
Aptamers are oligonucleotides that form secondary structures, giving them the ability to bind targeted molecules, including ions or small molecules, and, in our case, amanitin. BBa_K3593000 is an aptamer of α-amanitin named Best 1. It comes from one previous literature[1]. Its binding is tested through qPCR.
Sequence and features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterisation of this part
Experiment data
According to the state of art in aptamer binding[3], we design an experiment based on the principle of Gel Shift(i.e lower rate of motion in electrophoresis when aptamer is bound to the toxin due to a conformational change). By mixing excess amatoxin determined by the calculation of dissociation constant and aptamer in certain reaction times, we expect to see 2 bands representing the bound and inbound aptamer respectively. For examination of specificity, we also mix a random ssDNA pool where we select aptamer from and compare the band with the correct aptamer. For a high resolution, we used the TBE PAGE gel for DNA, 12%.
We also verify its binding affinity via a method called ELONA(enzyme-linked oligonucleotide assay), a method
from another literature[2], with a similar principle to ELISA.
References
1.Muszyńska K, Ostrowska D, Bartnicki F, et al. Selection and analysis of a DNA aptamer binding α-amanitin from Amanita phalloides. Acta Biochim Pol. 2017;64(3):401-406. 2.Han Q, Xia X, Jing L, et al. Selection and characterization of DNA aptamer specially targeting α-amanitin in wild mushrooms. SDRP J Food Sci Technol. 2018;3(6):497-508. 3.Muszyńska K, Ostrowska D, Bartnicki F, et al. Selection and analysis of a DNA aptamer binding α-amanitin from Amanita phalloides. Acta Biochim Pol. 2017;64(3):401-406. doi:10.18388/abp.2017_1615