Difference between revisions of "Part:BBa K3429011"
| Line 179: | Line 179: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ==Contribution: Team Aboa 2021== | |
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 14:02, 21 October 2021
Laccase CotA
Profile
| Name | CotA-Laccase (Spore coat protein A) |
| Base pairs | 1542 |
| Molecular weight | 58.51 kDa |
| Origin | Bacillus subtilis, synthetic |
| Properties | CotA laccase from B. subtilis is a copper dependant oxireductase with a broad substrate variety of polyphenolic substrates such as diclofenac and ABTS and possess a T1 copper for primary electron transfer to the substrate and a T2/3 Cu-cluster. |
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
CotA Docking
Ligand docking was performed on the laccase CotA (PDB 1GSK). Preparations of the ligand and protein were done as explained on our project wiki. For realization of the docking process we needed a working Rosetta xml script for our purpose. The script we worked out is shown in our Rosetta Guide. With this script, adjusted over many trials, we docked many thousand structures. As output we chose the Rosetta total score and the Rosetta interface score. We plotted these scores against the root-mean-square deviation of atomic positions (RMSD) from the ligand to allow a visible evaluation of the docked structures respectively poses[1]. The formula for calculation of the RMSD is shown below (formula 1).
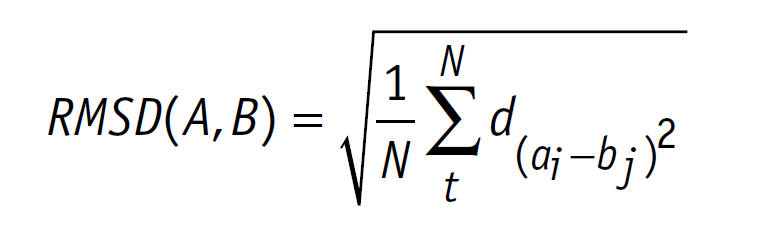
An interface score against RMSD plot from Diclofenac with CotA is shown in Figure 1. Based on these plots it was easier to identify low energy structures with good interactions with the enzyme. Ideally the data points of these plots should form a funnel like plot. The structures on the closing end are especially good structures. These structures are highlighted with by a red square in figure 1.

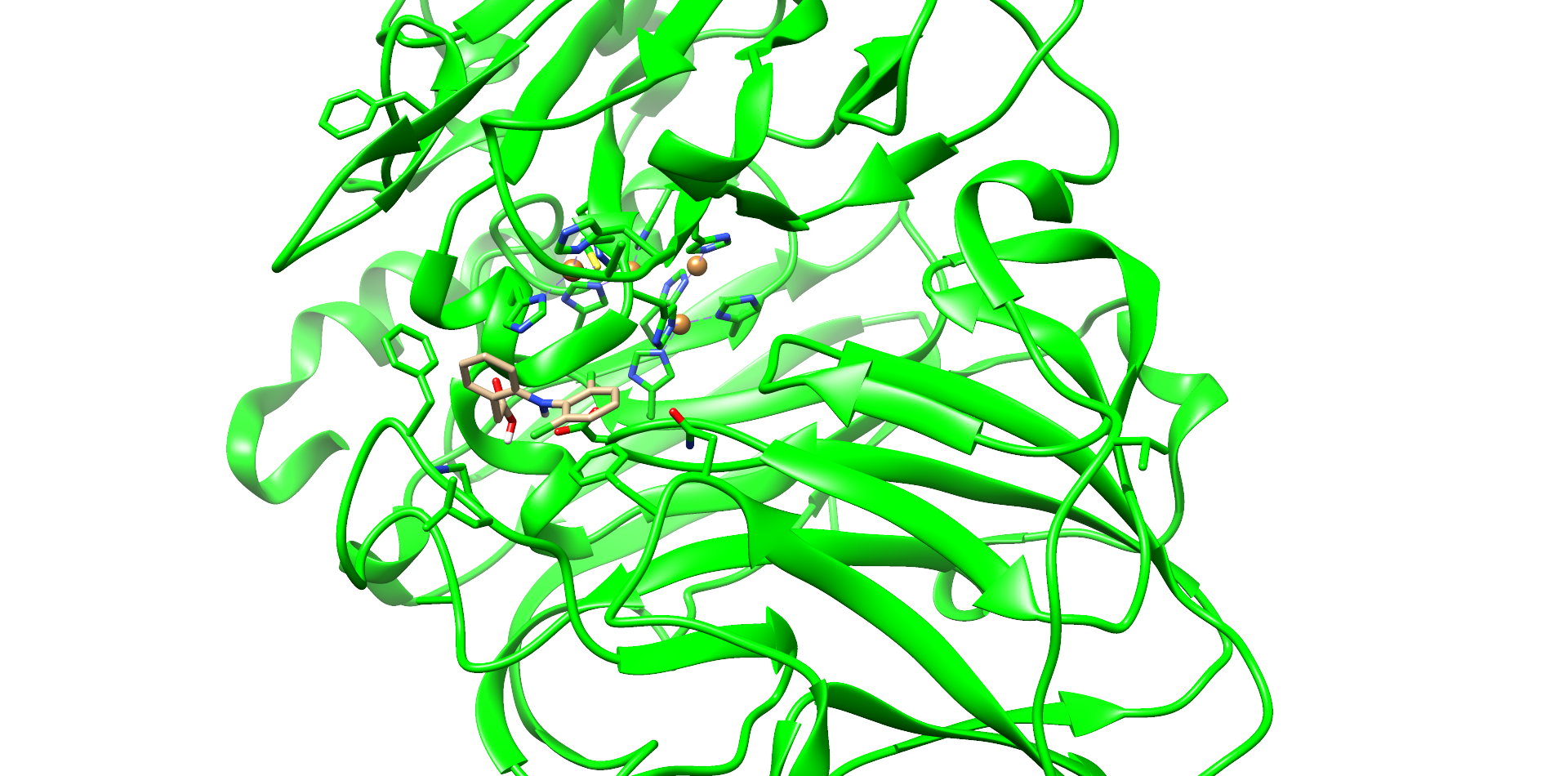
One of these diclofenac structures in CotA is shown in Figure 2. Interestingly, diclofenac was positioned here in a binding channel like structure and not directly near the active copper atom within the active site. This binding channel structure is flanked by two flexible loops of CotA. These loops could undergo a conformational change due to the binding of the substrate, which could hold the substrate in place or even bring it closer to the active site. The most dominant interaction in this model were with the residue’s threonine262 and threonine418. The threonine residues may interact with the secondary nitrogen and the carboxy group of diclofenac.
For comparison, the same docking procedure was performed with diclofenac and the laccase of T. versicolor. A structure with top scoring was also selected (Fig. 3). In this model diclofenac binds closer to the active site of the laccase of T. Versicolor. The substance also bound within a cavity of the enzyme and may interact by π-π-stacking with phenylalanine 163. Other interaction with the proline 163 and the carboxy group and aspartate 206 with the secondary nitrogen of diclofenac. The docking with the laccase from T. Versicolor showed more possible interactions and a better positioning of diclofenac near the active site. This assumption was supported by the interface and total score values of both docking experiments (table 2).
The following table 2 also shows an overview of the different scoring values of the top scored structures from different substrates like 4-Nonylphenol, Ibuprofen and Carbamazepin with CotA. The scoring values of all these substrates were relatively similar. Only the scoring values for the laccase from T. Versicolor were slightly better. But taking into account the positioning of the substance T. Versicolor is the superior laccase.
| Enzyme |
Substance |
Best Interface score |
Best Total score |
|---|---|---|---|
| CotA |
4-Nonylphenol |
-11.014 |
-1232.434 |
| Ibuprofen |
-12.170 |
-1232.533 |
|
| Carbamazepin |
-12.633 |
-1230.243 |
|
| Diclofenac |
-12.820 |
-1230.488 |
|
| T. Versicolor |
Diclofenac |
-13.266 |
-1528.821 |
[1] Steven A. Combs et al., Small-molecule ligand docking into comparative models with Rosetta, Nature Protocols, 2013, 8:1277–1298, doi:10.1038/nprot.2013.074