Difference between revisions of "Part:BBa K3512001"
EmiliaEmily (Talk | contribs) |
|||
| Line 10: | Line 10: | ||
[[Image:3D_ccdb.png|700px|thumb|center|]] | [[Image:3D_ccdb.png|700px|thumb|center|]] | ||
| + | |||
| + | ==Contribution: XHD-Wuhan-Pro-China 2021== | ||
| + | ===Literature 1=== | ||
| + | ===Toxicity and antitoxicity plate assays=== | ||
| + | To test the toxicity of the cloned CcdBO157 and CcdBF variants, the corresponding pBAD33-ccdB constructs were transformed in MG1655. The resulting transformants were plated on LB plates containing chloramphenicol with or without arabinose (1%). The CcdB variants were considered to be functional (toxic) when transformants were able to grow only in the absence of arabinose. To test the ability of the cloned CcdAO157 and CcdAF variants to counteract the toxicity of CcdBO157 and CcdBF, respectively, the corresponding pKK-ccdA constructs were transformed in MG1655 expressing the reference ccdBO157 orccdBF genes from the pBAD33 vector. The resulting transformants were plated on LB plates containing chloramphenicol and ampicillin with arabinose (1%). Basal expression of ccdA from the pTac promoter of pKK223-3 in MG1655 is sufficient to test the antitoxicity phenotype. The CcdA variants were considered to be functional when the toxicity of CcdBO157 or CcdBF protein was counteracted, i.e., when strains coexpressing a ccdA variant with the ccdBO157 or ccdBF reference genes were able to grow in the presence of arabinose while strains expressing only the ccdBO157 or ccdBF reference gene were not. | ||
| + | |||
| + | ===Variants of CcdB=== | ||
| + | Interestingly, the CcdB toxin proteins were much more diversified than the antitoxins. Among the 47 serogroups, 14 classes of alleles could be identified. Note that, as mentioned earlier, only one isolate for each serogroup was sequenced and tested (47 isolates). One class was composed of 4 isolates presenting sequence identical to the ccdBO157 gene of the O157:H7 EDL933 reference strain. The 2 most prevalent classes represented 13/47 and 8/47 isolates, and the corresponding alleles harbored either two variations (S10G and V28E) or one variation(S44I), respectively. The toxic activity of at least one representative CcdB protein of each class was tested using the toxicity plate assay and was comparable to that of the CcdB O157 protein. Four classes, together representing 7 isolates (7/47), contained from one to five variations (V28E,RH7-V28E,S10G-I26V-V28E,andRH7-S10G-I26V-V28E-I93L). At least one representative of each class was assayed for toxicity using the toxicity plate assay. Interestingly, expression of these variants led to cell killing, showing that the variations did not affect the toxic activity of the CcdB proteins. Ofthe7classesremaining, 1 is composed of 2 isolates (representing serogroups O138andO153) containing four variations in their ccdB gene (S10G-V28E-S44I-P54T). The CcdB O138 protein (from serogroup O138) was assayed for toxicity and surprisingly shown not to affect viability upon ectopic expression (Figure 5B). The comparison of the variations among the different classes suggests that the P54T mutation is responsible for abolishing the toxic activity of the CcdB O138 variant. The 6 last classes, representing 13/47 isolates, were composed of truncated proteins.Deletions of the carboxy-terminal region were caused by amber mutations at various locations (E41, R42, E63, and C84). Interestingly, all these truncated proteins contained several point variations that were also found in the full-length variants that were still toxic (S10G, I26V, V28E, and S44I) or not (R7H and P54T). Two of these classes (7/47 isolates) contained one extra variation (K62S), while another class (1/47 isolates) contained two more variations (R11S and R32S). One representative of 4 of these 6 classes was tested and was shown to be nontoxic, using the plate toxicity assay (data not shown). | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--9.jpg|750px|]] | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--10.jpg|750px|]] | ||
| + | |||
| + | ===Reference=== | ||
| + | Mine, N., Guglielmini, J., Wilbaux, M. and Van Melderen, L., 2009. The Decay of the Chromosomally Encoded ccdO157 Toxin–Antitoxin System in the Escherichia coli Species. Genetics, 181(4), pp.1557-1566. | ||
| + | |||
| + | ===Literature 2=== | ||
| + | ===Introduction=== | ||
| + | The F plasmid-carried bacterial toxin, the CcdB protein, is known to act on DNA gyrase in two different ways. CcdB poisons the gyrase-DNA complex, blocking the passage of polymerases and leading to double-strand breakage of the DNA. Alternatively, in cells that overexpress CcdB, the A subunit of DNA gyrase (GyrA) has been found as an inactive complex with CcdB. | ||
| + | |||
| + | In this work, we have reconstituted the inactive GyrA-CcdB complex in vitro, by denaturation-renaturation, and compared its properties with those of the complex made in vivo. We have reactivated the complex by the addition of CcdA or of CcdA41, a 41- residue C-terminal fragment of CcdA that has lost its autoregulatory properties but retains the ability to reverse the lethal effects of CcdB , and characterized the products of the reactivation procedure. We have also investigated the CcdB-resistant GyrA462 protein, the truncated GyrA 64- and 59-kDa N-terminal domain proteins, and the non-lethal CcdBo 15 mutant protein, all of which are unable to participate in CcdB-mediated cleavage of DNA, for their ability to replace wild-type GyrA or CcdB in the formation of the inactive complex. | ||
| + | |||
| + | ===Result=== | ||
| + | In Vivo Production and in Vitro Reconstitution of the GyrA-CcdB Inactive Complex | ||
| + | |||
| + | We have reconstituted an inactive GyrA-CcdB complex in vitro by denaturation and renaturation | ||
| + | of purified GyrA in the presence of excess CcdB. In order to directly compare the purification behavior and the physical and catalytic properties of this reconstituted complex with those of | ||
| + | the previously reported complex formed in vivo, we also purified the inactive GyrA-CcdB complex from a strain overproducing GyrA and CcdB proteins. When a portion of the renatured material was subjected to Superdex 200 gel filtration, two peaks were found (Fig. 1A). SDS-PAGE analysis revealed that the peak that eluted first contained both GyrA and CcdB; the second peak was free CcdB (Fig. 1B) | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--11.jpg|750px|]] | ||
| + | |||
| + | GyrA or GyrA462 was denatured and renatured in the presence of CcdB or CcdB°15. Each mixture was resolved by gel filtration on a Superdex 200 column eluted with 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 700 mM NaCl, and 10% (v/v) glycerol. (A) the elution profile shown is typical of that obtained with each of the above combinations. (B) complex formation was analyzed by subjecting both peaks from each profile to denaturing gel electrophoresis on a 4–20% polyacrylamide gradient gel run in Tris-glycine-SDS buffer and silver-stained. The control sample was a mixture of free GyrA and free CcdB proteins | ||
| + | |||
| + | The inactive GyrA-CcdB complexes produced in vitro and in vivo co-migrate in native gels at a position different from those of the free GyrA and CcdB proteins (data not shown) and exhibit a 90–95% inhibition of GyrB-dependent DNA supercoiling activity (Fig. 2). The strong resemblance between the in vivo and in vitro complexes indicates that the complex reconstituted in vitro is equivalent to that formed in vivo in CcdB-overproducing strains. No inactive complex and no reduction of GyrA-dependent supercoiling activity could be detected in in vitro control samples where the urea treatment was omitted. | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--12.jpg|750px|]] | ||
| + | |||
| + | The DNA supercoiling assay was performed. The 70-ml reactions contained the following proteins: GyrB (20 ng, lanes 2–9), GyrA (2 ng, lane2), GyrA-CcdB complex (10 ng, lanes 3 and 6; 50 ng, lanes 4 and 7; 100 ng, lane 8; 200 ng, lanes 5 and 9), and CcdA (200 ng, lanes 6–9). Reactions were incubated at 25 °C for 1 h, then terminated by addition of EDTA and SDS to final concentrations of 10 mM and 0.5%, respectively. Product DNA was analyzed on a 1% agarose gel run in TBE buffer. | ||
| + | |||
| + | Interestingly, complexes of CcdB with the truncated GyrA proteins appear to form spontaneously, since bands on native gels that correspond to these complexes were found in the control samples, which were not denatured with urea. An in vitro experiment with the GyrA 59-kDa protein and CcdB under nonchaotropic conditions confirmed this observation, and the highest yield of complex was obtained after a slow titration of CcdB into a GyrA59 solution (Fig. 4) | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--13.jpg|750px|]] | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--14.jpg|750px|]] | ||
| + | |||
| + | Complex formation between the GyrA 59-kDa N-terminal domain protein and CcdB under non-chaotropic conditions. | ||
| + | |||
| + | (A) after a slow titration of a molar excess of CcdB into a GyrA59 solution in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, the sample was resolved on a Superdex 75 HR (10/30) gel filtration column in the same buffer. | ||
| + | |||
| + | (B) protein peak fractions were analyzed by SDS-PAGE. | ||
| + | |||
| + | In Vitro Reactivation of the Reconstituted GyrA-CcdB Complex by CcdA and CcdA41 | ||
| + | In the presence of CcdA, the GyrB-dependent supercoiling activity was entirely recovered. CcdA can also restore ATP- and GyrB-dependent DNA relaxing activity to the inactive CcdB complexes in which the truncated GyrA 64- and 59 kDa N-terminal proteins are substituted for the full-length GyrA protein (Fig. 3). | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--15.jpg|750px|]] | ||
| + | |||
| + | Inactivation of GyrA N-terminal 64- and 59-kDa domains by CcdB; reactivation by CcdA. | ||
| + | |||
| + | DNA relaxation assay buffer conditions were the same as the conditions of the cleavage reactions described in the legend to Fig. 7. Truncated GyrA proteins were first denatured and renatured in the absence or presence of CcdB as described under “Experimental Procedures.” GyrA64(2CcdB) (110 ng, lane 2; 220 ng, lane 3), GyrA64(1CcdB) (110 ng, lanes 4 and 6; 220 ng, lanes 5 and 7), GyrA59 (2CcdB) (27 ng, lane 9; 54 ng, lane 10), GyrA59(1CcdB) (27 ng, lanes 11 and 13; 54 ng, lanes 12 and 14), and CcdA (2.8 mg, lanes 6, 7, 13, 14, and 15) were incubated at 30 °C for 30 min in 70 ml of reaction buffer lacking ATP. GyrB protein (50 ng, lanes 9, 11, and 13; 100 ng, lanes 2, 4, 6, 10, 12, and 14; 200 ng, lanes 3, 5, and 7), negatively supercoiled pBR322 DNA (0.7 mg, lanes 1–7 and 9–15), relaxed pBR322 DNA (0.7 mg, lane 8), and ATP (1.4 mM, lanes 1–15) were then added, and incubation was continued at 30 °C for 2 h. After removal of protein, the samples were analyzed on an 0.8% agarose gel run in TBE buffer. | ||
| + | |||
| + | After a brief incubation of CcdA or CcdA41 with the GyrA-CcdB complex, the mixtures were resolved on Superdex 200 gel filtration columns. The gel filtration profiles show that when the GyrA-CcdB complex was run alone, only one peak, corresponding to this GyrA-CcdB complex, was obtained. However, when either CcdA or CcdA41 was added in excess over CcdB to the complex, three peaks were obtained (Fig. 5A). Each peak was analyzed on a polyacrylamide denaturing gel (Fig. 5B). The first peak corresponds to free GyrA and contains no CcdB; the second peak contains the newly formed CcdB-CcdA or CcdB-CcdA41 complex; the third | ||
| + | peak corresponds to the free CcdA or CcdA41. | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--16.jpg|750px|]] | ||
| + | |||
| + | Protein species present after reactivation of the reconstituted GyrA-CcdB complex | ||
| + | |||
| + | After preincubation of the reconstituted GyrA-CcdB complex (20 mg) with CcdA (10 mg) or CcdA41 (10 mg) for 5 min at room temperature, the mixture was passed through a Superdex 200 gel filtration column equilibrated with 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 700 mM NaCl, and 10% (v/v) glycerol, at a flow rate of 50 ml/min. (A) elution volume positions of the proteins are indicated by the arrows on the gel filtration profiles. Profile 1 corresponds to the GyrA-CcdB complex before reactivation. Profiles 2 and 3 correspond to the GyrA-CcdB complex reactivated with CcdA or CcdA41, respectively. (B) each peak in profiles 1 and 2 was analyzed on a 4–20% polyacrylamide gradient gel run in Trisglycine-SDS buffer and silver-stained. | ||
| + | |||
| + | A similar result was obtained when the CcdB-GyrA 59-kDa protein complex was formed under non-denaturing conditions on a GyrA59-Sepharose 4B column, followed by elution of the CcdB from the complex with an excess of CcdA. Upon fractionation of the eluted material by Superdex 75 gel filtration, we found one peak corresponding to the tetrameric complex (CcdA)2(CcdB)2 with a molecular weight of 41,000, and a second peak corresponding to (CcdA)2 with a molecular weight of 20,000 (Fig. 6). | ||
| + | |||
| + | [[File:T-- XHD-Wuhan-Pro-China--17.jpg|750px|]] | ||
| + | |||
| + | Complex formation between CcdA and CcdB on a GyrA59 column. | ||
| + | |||
| + | A 1.0-ml GyrA59-Sepharose 4B FPLC column was charged with CcdB and subsequently eluted with CcdA. (A) the eluted CcdA-CcdB complex was further fractionated on a Superdex 75 HR (10/30) column in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl. ( B) peak fractions were analyzed by SDS-PAGE. | ||
| + | |||
| + | These observations demonstrate that CcdB is quickly and completely extracted from a GyrA-CcdB complex by CcdA and by CcdA41 and that new CcdB-CcdA or CcdB-CcdA41 complexes are formed. Thus, the affinity of CcdB for CcdA or CcdA41 is apparently greater than its affinity for GyrA. | ||
| + | |||
| + | Mutations in the GyrA and CcdB Proteins Which Prevent Poisoning of the Cleavable Complex Also Prevent GyrA Inactivation | ||
| + | CcdB has been shown to trap DNA gyrase in a cleavable complex such that, on addition of SDS or alkali, double-strand cleavage of DNA is observed. The GyrA462 mutant protein, which confers | ||
| + | resistance to CcdB in vivo, and CcdBo15, a non-killer CcdB protein , were used to perform CcdB-mediated cleavage. reactions. GyrA462, in the presence of GyrB, is unable to promote any CcdB-mediated DNA cleavage (Ref. 27; Fig. 7). Similarly, when CcdBo15 was assayed | ||
| + | for in vitro DNA cleavage in presence of GyrA and GyrB, no DNA cleavage was observed (Fig. 7). These mutant proteins were then tested for their ability to form inactive complexes | ||
| + | (GyrA462-CcdB, GyrA-CcdBo15, or GyrA462-CcdBo15) in vitro. | ||
| + | |||
| + | After denaturation and renaturation, the protein solutions were resolved on Superdex 200 gel filtration columns (see Fig. 1A). Fractions from each peak were evaluated by denaturing gel electrophoresis. The complex was formed only when the wild type GyrA and CcdB proteins were used together; with all the other combinations, no complex was formed (Fig. 1B) and no inhibition of GyrB-dependent supercoiling was found (data not shown). Attempts to produce these complexes in cells overexpressing CcdBo15 with wild-type GyrA or wild-type CcdB with GyrA462 were also unsuccessful, as no complexes could be detected. From these results, we conclude that the protein interactions between CcdB and gyrase in the cleavable DNA complex and between CcdB and GyrA in the inactive GyrA-CcdB complex share essential site. | ||
| + | |||
| + | ===Reference=== | ||
| + | Bahassi, E., O'Dea, M., Allali, N., Messens, J., Gellert, M. and Couturier, M., 1999. Interactions of CcdB with DNA Gyrase. Journal of Biological Chemistry, 274(16), pp.10936-10944. | ||
| + | |||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 14:36, 19 October 2021
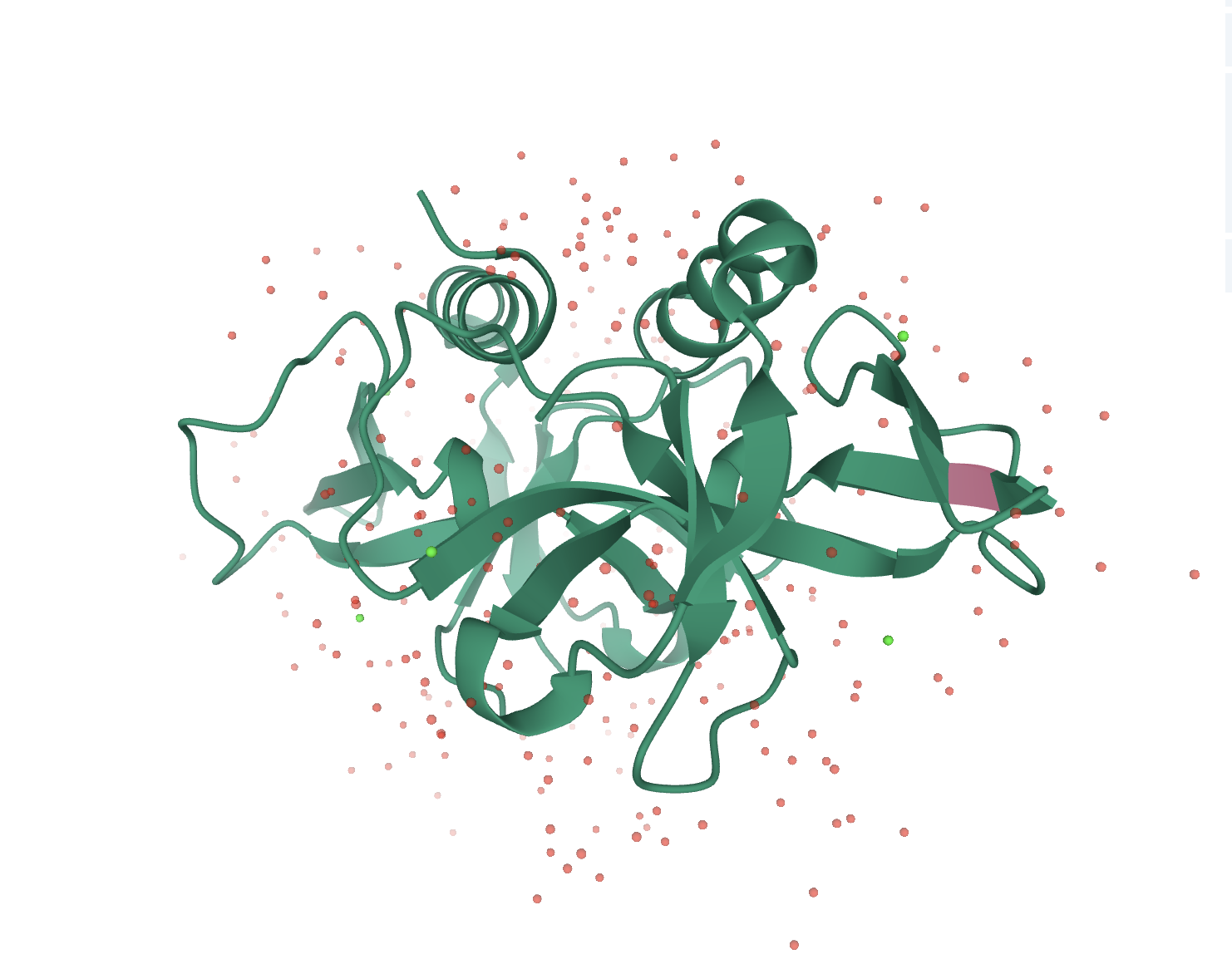
ccdB Toxin
The CcdB toxin is part of the CcdA-CcdB system. The target of CcdB is the GyrA subunit of DNA gyrase, an essential type II topoisomerase in Escherichia coli. Gyrase alters DNA topology by effecting a transient double-strand break in the DNA backbone, passing the double helix through the gate and resealing the gaps. The CcdB toxin acts by trapping DNA gyrase in a cleaved complex with the gyrase A subunit covalently closed to the cleaved DNA, causing DNA breakage and cell death in a way closely related to quinolones antibiotics.
In absence of the antitoxin CcdA, the CcdB toxin traps DNA-gyrase cleavable complexes, inducing breaks into DNA and cell death.
3D View
Contribution: XHD-Wuhan-Pro-China 2021
Literature 1
Toxicity and antitoxicity plate assays
To test the toxicity of the cloned CcdBO157 and CcdBF variants, the corresponding pBAD33-ccdB constructs were transformed in MG1655. The resulting transformants were plated on LB plates containing chloramphenicol with or without arabinose (1%). The CcdB variants were considered to be functional (toxic) when transformants were able to grow only in the absence of arabinose. To test the ability of the cloned CcdAO157 and CcdAF variants to counteract the toxicity of CcdBO157 and CcdBF, respectively, the corresponding pKK-ccdA constructs were transformed in MG1655 expressing the reference ccdBO157 orccdBF genes from the pBAD33 vector. The resulting transformants were plated on LB plates containing chloramphenicol and ampicillin with arabinose (1%). Basal expression of ccdA from the pTac promoter of pKK223-3 in MG1655 is sufficient to test the antitoxicity phenotype. The CcdA variants were considered to be functional when the toxicity of CcdBO157 or CcdBF protein was counteracted, i.e., when strains coexpressing a ccdA variant with the ccdBO157 or ccdBF reference genes were able to grow in the presence of arabinose while strains expressing only the ccdBO157 or ccdBF reference gene were not.
Variants of CcdB
Interestingly, the CcdB toxin proteins were much more diversified than the antitoxins. Among the 47 serogroups, 14 classes of alleles could be identified. Note that, as mentioned earlier, only one isolate for each serogroup was sequenced and tested (47 isolates). One class was composed of 4 isolates presenting sequence identical to the ccdBO157 gene of the O157:H7 EDL933 reference strain. The 2 most prevalent classes represented 13/47 and 8/47 isolates, and the corresponding alleles harbored either two variations (S10G and V28E) or one variation(S44I), respectively. The toxic activity of at least one representative CcdB protein of each class was tested using the toxicity plate assay and was comparable to that of the CcdB O157 protein. Four classes, together representing 7 isolates (7/47), contained from one to five variations (V28E,RH7-V28E,S10G-I26V-V28E,andRH7-S10G-I26V-V28E-I93L). At least one representative of each class was assayed for toxicity using the toxicity plate assay. Interestingly, expression of these variants led to cell killing, showing that the variations did not affect the toxic activity of the CcdB proteins. Ofthe7classesremaining, 1 is composed of 2 isolates (representing serogroups O138andO153) containing four variations in their ccdB gene (S10G-V28E-S44I-P54T). The CcdB O138 protein (from serogroup O138) was assayed for toxicity and surprisingly shown not to affect viability upon ectopic expression (Figure 5B). The comparison of the variations among the different classes suggests that the P54T mutation is responsible for abolishing the toxic activity of the CcdB O138 variant. The 6 last classes, representing 13/47 isolates, were composed of truncated proteins.Deletions of the carboxy-terminal region were caused by amber mutations at various locations (E41, R42, E63, and C84). Interestingly, all these truncated proteins contained several point variations that were also found in the full-length variants that were still toxic (S10G, I26V, V28E, and S44I) or not (R7H and P54T). Two of these classes (7/47 isolates) contained one extra variation (K62S), while another class (1/47 isolates) contained two more variations (R11S and R32S). One representative of 4 of these 6 classes was tested and was shown to be nontoxic, using the plate toxicity assay (data not shown).
Reference
Mine, N., Guglielmini, J., Wilbaux, M. and Van Melderen, L., 2009. The Decay of the Chromosomally Encoded ccdO157 Toxin–Antitoxin System in the Escherichia coli Species. Genetics, 181(4), pp.1557-1566.
Literature 2
Introduction
The F plasmid-carried bacterial toxin, the CcdB protein, is known to act on DNA gyrase in two different ways. CcdB poisons the gyrase-DNA complex, blocking the passage of polymerases and leading to double-strand breakage of the DNA. Alternatively, in cells that overexpress CcdB, the A subunit of DNA gyrase (GyrA) has been found as an inactive complex with CcdB.
In this work, we have reconstituted the inactive GyrA-CcdB complex in vitro, by denaturation-renaturation, and compared its properties with those of the complex made in vivo. We have reactivated the complex by the addition of CcdA or of CcdA41, a 41- residue C-terminal fragment of CcdA that has lost its autoregulatory properties but retains the ability to reverse the lethal effects of CcdB , and characterized the products of the reactivation procedure. We have also investigated the CcdB-resistant GyrA462 protein, the truncated GyrA 64- and 59-kDa N-terminal domain proteins, and the non-lethal CcdBo 15 mutant protein, all of which are unable to participate in CcdB-mediated cleavage of DNA, for their ability to replace wild-type GyrA or CcdB in the formation of the inactive complex.
Result
In Vivo Production and in Vitro Reconstitution of the GyrA-CcdB Inactive Complex
We have reconstituted an inactive GyrA-CcdB complex in vitro by denaturation and renaturation of purified GyrA in the presence of excess CcdB. In order to directly compare the purification behavior and the physical and catalytic properties of this reconstituted complex with those of the previously reported complex formed in vivo, we also purified the inactive GyrA-CcdB complex from a strain overproducing GyrA and CcdB proteins. When a portion of the renatured material was subjected to Superdex 200 gel filtration, two peaks were found (Fig. 1A). SDS-PAGE analysis revealed that the peak that eluted first contained both GyrA and CcdB; the second peak was free CcdB (Fig. 1B)
GyrA or GyrA462 was denatured and renatured in the presence of CcdB or CcdB°15. Each mixture was resolved by gel filtration on a Superdex 200 column eluted with 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 700 mM NaCl, and 10% (v/v) glycerol. (A) the elution profile shown is typical of that obtained with each of the above combinations. (B) complex formation was analyzed by subjecting both peaks from each profile to denaturing gel electrophoresis on a 4–20% polyacrylamide gradient gel run in Tris-glycine-SDS buffer and silver-stained. The control sample was a mixture of free GyrA and free CcdB proteins
The inactive GyrA-CcdB complexes produced in vitro and in vivo co-migrate in native gels at a position different from those of the free GyrA and CcdB proteins (data not shown) and exhibit a 90–95% inhibition of GyrB-dependent DNA supercoiling activity (Fig. 2). The strong resemblance between the in vivo and in vitro complexes indicates that the complex reconstituted in vitro is equivalent to that formed in vivo in CcdB-overproducing strains. No inactive complex and no reduction of GyrA-dependent supercoiling activity could be detected in in vitro control samples where the urea treatment was omitted.
The DNA supercoiling assay was performed. The 70-ml reactions contained the following proteins: GyrB (20 ng, lanes 2–9), GyrA (2 ng, lane2), GyrA-CcdB complex (10 ng, lanes 3 and 6; 50 ng, lanes 4 and 7; 100 ng, lane 8; 200 ng, lanes 5 and 9), and CcdA (200 ng, lanes 6–9). Reactions were incubated at 25 °C for 1 h, then terminated by addition of EDTA and SDS to final concentrations of 10 mM and 0.5%, respectively. Product DNA was analyzed on a 1% agarose gel run in TBE buffer.
Interestingly, complexes of CcdB with the truncated GyrA proteins appear to form spontaneously, since bands on native gels that correspond to these complexes were found in the control samples, which were not denatured with urea. An in vitro experiment with the GyrA 59-kDa protein and CcdB under nonchaotropic conditions confirmed this observation, and the highest yield of complex was obtained after a slow titration of CcdB into a GyrA59 solution (Fig. 4)
Complex formation between the GyrA 59-kDa N-terminal domain protein and CcdB under non-chaotropic conditions.
(A) after a slow titration of a molar excess of CcdB into a GyrA59 solution in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, the sample was resolved on a Superdex 75 HR (10/30) gel filtration column in the same buffer.
(B) protein peak fractions were analyzed by SDS-PAGE.
In Vitro Reactivation of the Reconstituted GyrA-CcdB Complex by CcdA and CcdA41 In the presence of CcdA, the GyrB-dependent supercoiling activity was entirely recovered. CcdA can also restore ATP- and GyrB-dependent DNA relaxing activity to the inactive CcdB complexes in which the truncated GyrA 64- and 59 kDa N-terminal proteins are substituted for the full-length GyrA protein (Fig. 3).
Inactivation of GyrA N-terminal 64- and 59-kDa domains by CcdB; reactivation by CcdA.
DNA relaxation assay buffer conditions were the same as the conditions of the cleavage reactions described in the legend to Fig. 7. Truncated GyrA proteins were first denatured and renatured in the absence or presence of CcdB as described under “Experimental Procedures.” GyrA64(2CcdB) (110 ng, lane 2; 220 ng, lane 3), GyrA64(1CcdB) (110 ng, lanes 4 and 6; 220 ng, lanes 5 and 7), GyrA59 (2CcdB) (27 ng, lane 9; 54 ng, lane 10), GyrA59(1CcdB) (27 ng, lanes 11 and 13; 54 ng, lanes 12 and 14), and CcdA (2.8 mg, lanes 6, 7, 13, 14, and 15) were incubated at 30 °C for 30 min in 70 ml of reaction buffer lacking ATP. GyrB protein (50 ng, lanes 9, 11, and 13; 100 ng, lanes 2, 4, 6, 10, 12, and 14; 200 ng, lanes 3, 5, and 7), negatively supercoiled pBR322 DNA (0.7 mg, lanes 1–7 and 9–15), relaxed pBR322 DNA (0.7 mg, lane 8), and ATP (1.4 mM, lanes 1–15) were then added, and incubation was continued at 30 °C for 2 h. After removal of protein, the samples were analyzed on an 0.8% agarose gel run in TBE buffer.
After a brief incubation of CcdA or CcdA41 with the GyrA-CcdB complex, the mixtures were resolved on Superdex 200 gel filtration columns. The gel filtration profiles show that when the GyrA-CcdB complex was run alone, only one peak, corresponding to this GyrA-CcdB complex, was obtained. However, when either CcdA or CcdA41 was added in excess over CcdB to the complex, three peaks were obtained (Fig. 5A). Each peak was analyzed on a polyacrylamide denaturing gel (Fig. 5B). The first peak corresponds to free GyrA and contains no CcdB; the second peak contains the newly formed CcdB-CcdA or CcdB-CcdA41 complex; the third peak corresponds to the free CcdA or CcdA41.
Protein species present after reactivation of the reconstituted GyrA-CcdB complex
After preincubation of the reconstituted GyrA-CcdB complex (20 mg) with CcdA (10 mg) or CcdA41 (10 mg) for 5 min at room temperature, the mixture was passed through a Superdex 200 gel filtration column equilibrated with 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 700 mM NaCl, and 10% (v/v) glycerol, at a flow rate of 50 ml/min. (A) elution volume positions of the proteins are indicated by the arrows on the gel filtration profiles. Profile 1 corresponds to the GyrA-CcdB complex before reactivation. Profiles 2 and 3 correspond to the GyrA-CcdB complex reactivated with CcdA or CcdA41, respectively. (B) each peak in profiles 1 and 2 was analyzed on a 4–20% polyacrylamide gradient gel run in Trisglycine-SDS buffer and silver-stained.
A similar result was obtained when the CcdB-GyrA 59-kDa protein complex was formed under non-denaturing conditions on a GyrA59-Sepharose 4B column, followed by elution of the CcdB from the complex with an excess of CcdA. Upon fractionation of the eluted material by Superdex 75 gel filtration, we found one peak corresponding to the tetrameric complex (CcdA)2(CcdB)2 with a molecular weight of 41,000, and a second peak corresponding to (CcdA)2 with a molecular weight of 20,000 (Fig. 6).
Complex formation between CcdA and CcdB on a GyrA59 column.
A 1.0-ml GyrA59-Sepharose 4B FPLC column was charged with CcdB and subsequently eluted with CcdA. (A) the eluted CcdA-CcdB complex was further fractionated on a Superdex 75 HR (10/30) column in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl. ( B) peak fractions were analyzed by SDS-PAGE.
These observations demonstrate that CcdB is quickly and completely extracted from a GyrA-CcdB complex by CcdA and by CcdA41 and that new CcdB-CcdA or CcdB-CcdA41 complexes are formed. Thus, the affinity of CcdB for CcdA or CcdA41 is apparently greater than its affinity for GyrA.
Mutations in the GyrA and CcdB Proteins Which Prevent Poisoning of the Cleavable Complex Also Prevent GyrA Inactivation CcdB has been shown to trap DNA gyrase in a cleavable complex such that, on addition of SDS or alkali, double-strand cleavage of DNA is observed. The GyrA462 mutant protein, which confers resistance to CcdB in vivo, and CcdBo15, a non-killer CcdB protein , were used to perform CcdB-mediated cleavage. reactions. GyrA462, in the presence of GyrB, is unable to promote any CcdB-mediated DNA cleavage (Ref. 27; Fig. 7). Similarly, when CcdBo15 was assayed for in vitro DNA cleavage in presence of GyrA and GyrB, no DNA cleavage was observed (Fig. 7). These mutant proteins were then tested for their ability to form inactive complexes (GyrA462-CcdB, GyrA-CcdBo15, or GyrA462-CcdBo15) in vitro.
After denaturation and renaturation, the protein solutions were resolved on Superdex 200 gel filtration columns (see Fig. 1A). Fractions from each peak were evaluated by denaturing gel electrophoresis. The complex was formed only when the wild type GyrA and CcdB proteins were used together; with all the other combinations, no complex was formed (Fig. 1B) and no inhibition of GyrB-dependent supercoiling was found (data not shown). Attempts to produce these complexes in cells overexpressing CcdBo15 with wild-type GyrA or wild-type CcdB with GyrA462 were also unsuccessful, as no complexes could be detected. From these results, we conclude that the protein interactions between CcdB and gyrase in the cleavable DNA complex and between CcdB and GyrA in the inactive GyrA-CcdB complex share essential site.
Reference
Bahassi, E., O'Dea, M., Allali, N., Messens, J., Gellert, M. and Couturier, M., 1999. Interactions of CcdB with DNA Gyrase. Journal of Biological Chemistry, 274(16), pp.10936-10944.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 216