Difference between revisions of "Part:BBa K3407006"
| Line 33: | Line 33: | ||
For each short hairpin, a pair of complementary primers were annealed to form a DNA template for transcription (Figure 2B). The DNA template was designed to possess a T7 promoter followed by a 27nt inverted repeat sequence taken from the eGFP gene (nt 78 to 105), and linked together by the 9nt sequence containing the target of Fox-1 RBD. On the 3’ termini, two GG were added to produce the desired overhang (<html><a href="https://parts.igem.org/Part:BBa_K3407002" target="_blank"><b>BBa_K3407022</b></a><html>). This leads to a successful transcription without the need to use a terminator (Figure 2A). | For each short hairpin, a pair of complementary primers were annealed to form a DNA template for transcription (Figure 2B). The DNA template was designed to possess a T7 promoter followed by a 27nt inverted repeat sequence taken from the eGFP gene (nt 78 to 105), and linked together by the 9nt sequence containing the target of Fox-1 RBD. On the 3’ termini, two GG were added to produce the desired overhang (<html><a href="https://parts.igem.org/Part:BBa_K3407002" target="_blank"><b>BBa_K3407022</b></a><html>). This leads to a successful transcription without the need to use a terminator (Figure 2A). | ||
| + | <div><ul> | ||
| + | <center> | ||
| + | <li style="display: inline-block;"> [[File:T--TUDelft--RiboMAX.jpeg|thumb|none|800px|<b>Figure 2:</b> Schematic representation of the shRNA in vitro production using T7 RiboMAX kit from Promega. Primer pairs are annealed in the T4 ligase buffer by heating at 95ºC for 5 minutes and left cooling at room temperature for 30 minutes. Once template is annealed, T7 RNA polymerase is added to transcribe the DNA template at 42ºC for 2 hours. shRNAs self assemble as soon as they are produced. (B) DNA template design for in vitro shRNA production. T7 promoter followed by eGFP target sequences with inverted repeats are linked by the loop region, and finally ends with a GG overhang.]] </li> | ||
| + | </center> | ||
| + | </ul></div> | ||
| + | |||
| + | ==a== | ||
| + | shRNA (<html><a href="https://parts.igem.org/Part:BBa_K3407022" target="_blank"><b>BBa_K3407022</b></a><html>), shRNA* (<html><a href="https://parts.igem.org/Part:BBa_K3407023" target="_blank"><b>BBa_K3407023</b></a><html>) | ||
===References=== | ===References=== | ||
Revision as of 12:14, 26 October 2020
Short hairpin RNA (shRNA): a dmDicer-2 substrate and potential trigger of RNAi.
Usage and Biology
shRNAs are single RNA molecules with a strong, self-assembling secondary structure and a loop. In the scientific community, RNA interference (RNAi) terminology can be confusing by referring to different molecules with the same name. The diverse nature of RNA structures makes it difficult to reflect its heterogeneity while keeping a minimum number of terms. Double-stranded RNA (dsRNA) is commonly referred to as complementary sequences of RNA, that hybridise to form a fragment of dsRNA of about 200-600bp [1][2][3][4][5]; others use the term to name shRNAs [6], while some refer to shRNAs as such [7][8]. The reality is, many structures of single-stranded RNAs (ssRNAs) can hybridise with others or themselves to become fully double-stranded, or reserve only a specific part of the whole to form dsRNA.
RNAi is triggered when dsRNA is present in the cytoplasm of most eukaryotes. Dicer is the enzyme in charge of recognising and cleaving dsRNA into either micro RNA (miRNA), or small interfering RNA (siRNA) [9]. In eukaryotes other than plants, siRNA arise from the processing of dsRNA without mismatches, while miRNA are mainly thought to come from mismatched dsRNA processing. In insects, miRNAs are a product of Dicer-1 processing, while siRNA are generated by Dicer-2 [10], at least the latter in a ATP-independent manner [11]. One strand of these small RNAs is selected and loaded in RNA Induced Silencing Complex (RISC), to direct post-transcriptional gene silencing. It is attained by interfering with mRNAs that show a certain degree of complementarity with the selected strand from miRNAs, or by cleaving the mRNA if it shows 100% complementarity with the selected strand from siRNA [12]. This statements remains partially true, as crossovers between the two pathways have been shown to happen, as siRNA can behave like miRNA by interfering with mRNAs with only a certain degree of complementarity, corresponding to a 100% match only with the “seed region” (2-8 nt) of the loaded strand in RISC [13].
RNA hairpins can take both pathways, leading to either siRNA or miRNA. To produce miRNA, a transcript called pri-miRNA is processed by Drosha-DGCR8 complex into pre-miRNA in the nucleus, to later be exported to the cytoplasm by Exportin-5 [14]. To do so, this latter protein needs to recognise the loop of the pre-miRNA and its overhang [15]. Next, pre-miRNA is further processed by Dicer to produce miRNAs [16][17]. On the other hand, when short RNA hairpins do not show any mismatches in their dsRNA sequence, they can be processed by Dicer-2 to produce siRNA [18], as depicted in Figure 1. Interestingly, shRNA can bypass Dicer processing to perform siRNA-like silencing by directly loading into RISC [19]. Therefore, shRNAs are valuable candidates to mediate specific gene silencing through siRNA pathway.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Experimental results
shRNA can be transcribed with T7 RNA polymerase
shRNA are produced in vitro using the T7 RiboMAX RNAi express kit from Promega, which makes use of the viral T7 RNA polymerase.
-
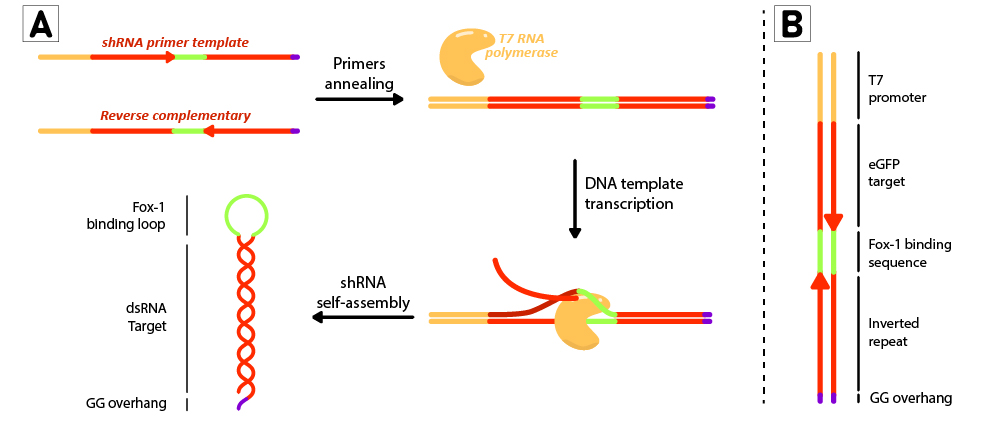 Figure 2: Schematic representation of the shRNA in vitro production using T7 RiboMAX kit from Promega. Primer pairs are annealed in the T4 ligase buffer by heating at 95ºC for 5 minutes and left cooling at room temperature for 30 minutes. Once template is annealed, T7 RNA polymerase is added to transcribe the DNA template at 42ºC for 2 hours. shRNAs self assemble as soon as they are produced. (B) DNA template design for in vitro shRNA production. T7 promoter followed by eGFP target sequences with inverted repeats are linked by the loop region, and finally ends with a GG overhang.
Figure 2: Schematic representation of the shRNA in vitro production using T7 RiboMAX kit from Promega. Primer pairs are annealed in the T4 ligase buffer by heating at 95ºC for 5 minutes and left cooling at room temperature for 30 minutes. Once template is annealed, T7 RNA polymerase is added to transcribe the DNA template at 42ºC for 2 hours. shRNAs self assemble as soon as they are produced. (B) DNA template design for in vitro shRNA production. T7 promoter followed by eGFP target sequences with inverted repeats are linked by the loop region, and finally ends with a GG overhang.
For each short hairpin, a pair of complementary primers were annealed to form a DNA template for transcription (Figure 2B). The DNA template was designed to possess a T7 promoter followed by a 27nt inverted repeat sequence taken from the eGFP gene (nt 78 to 105), and linked together by the 9nt sequence containing the target of Fox-1 RBD. On the 3’ termini, two GG were added to produce the desired overhang (BBa_K3407022). This leads to a successful transcription without the need to use a terminator (Figure 2A).
- [[File:T--TUDelft--RiboMAX.jpeg|thumb|none|800px|Figure 2: Schematic representation of the shRNA in vitro production using T7 RiboMAX kit from Promega. Primer pairs are annealed in the T4 ligase buffer by heating at 95ºC for 5 minutes and left cooling at room temperature for 30 minutes. Once template is annealed, T7 RNA polymerase is added to transcribe the DNA template at 42ºC for 2 hours. shRNAs self assemble as soon as they are produced. (B) DNA template design for in vitro shRNA production. T7 promoter followed by eGFP target sequences with inverted repeats are linked by the loop region, and finally ends with a GG overhang.]]
- Lenaerts, C., Palmans, J., Marchal, E., Verdonck, R. and Vanden Broeck, J., 2020. Role Of The Venus Kinase Receptor In The Female Reproductive Physiology Of The Desert Locust, Schistocerca Gregaria. Sci Rep 7.
- Lenaerts, C., Marchal, E., Peeters, P. and Vanden Broeck, J., 2020. The Ecdysone Receptor Complex Is Essential For The Reproductive Success In The Female Desert Locust, Schistocerca Gregaria. 9
- Marchal, E., Badisco, L., Verlinden, H., Vandersmissen, T., Van Soest, S., Van Wielendaele, P. and Vanden Broeck, J., 2020. Role Of The Halloween Genes, Spook And Phantom In Ecdysteroidogenesis In The Desert Locust, Schistocerca Gregaria.
- Vatanparast, M. and Kim, Y., 2020. Optimization Of Recombinant Bacteria Expressing Dsrna To Enhance Insecticidal Activity Against A Lepidopteran Insect, Spodoptera Exigua.
- Adeyinka, O., Tabassum, B., Nasir, I., Yousaf, I., Sajid, I., Shehzad, K., Batcho, A. and Husnain, T., 2020. Identification And Validation Of Potential Reference Gene For Effective Dsrna Knockdown Analysis In Chilo Partellus.
- Ma, Z., Zhou, H., Wei, Y., Yan, S. and Shen, J., 2020. A Novel Plasmid– Escherichia Coli System Produces Large Batch Dsrnas For Insect Gene Silencing.
- Yuka M., Rina K., Hiroyuki S., Tamaki E., and Takashi O., 2011. Photosensitizing Carrier Proteins for Photoinducible RNA Interference.
- Soler Canton A., Danelon C., Dogterom A., 2020. Synthetic Biology Meets Liposome-Based Drug Delivery. [online] Repository.tudelft.nl.
- Zhang, H., Kolb, F., Jaskiewicz, L., Westhof, E. and Filipowicz, W., 2020. Single Processing Center Models For Human Dicer And Bacterial Rnase III.
- Young S., Kenji N., John W., Kevin K., Zhengying H., Erik J., Richard W., 2004. Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA Silencing Pathways.
- Sinha, N., Trettin, K., Aruscavage, P. and Bass, B., 2020. Drosophila Dicer-2 Cleavage Is Mediated By Helicase- And Dsrna Termini-Dependent States That Are Modulated By Loquacious-PD.
- Lam, J., Chow, M., Zhang, Y. and Leung, S., 2020. Sirna Versus Mirna As Therapeutics For Gene Silencing. 9
- Burchard, J., Jackson, A., Malkov, V., Needham, R., Tan, Y., Bartz, S., Dai, H., Sachs, A. and Linsley, P., 2020. Microrna-Like Off-Target Transcript Regulation By Sirnas Is Species Specific.
- Bofill-De Ros, X. and Gu, S., 2020. Guidelines For The Optimal Design Of Mirna-Based Shrnas.
- Wang, X., Xu, X., Ma, Z., Huo, Y., Xiao, Z., Li, Y. and Wang, Y., 2020. Dynamic Mechanisms For Pre-Mirna Binding And Export By Exportin-5.
- Bofill-De Ros, X. and Gu, S., 2020. Guidelines For The Optimal Design Of Mirna-Based Shrnas.
- Lam, J., Chow, M., Zhang, Y. and Leung, S., 2020. Sirna Versus Mirna As Therapeutics For Gene Silencing.
- Sioud, M., 2020. What Are The Key Targeted Delivery Technologies Of Sirna Now?.
- Liu, Y., Schopman, N. and Berkhout, B., 2020. Dicer-Independent Processing Of Short Hairpin Rnas.

