Difference between revisions of "Part:BBa K3425005"
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K3425005 short</partinfo> | <partinfo>BBa_K3425005 short</partinfo> | ||
| + | This, pSB3K01, part is a medium copy number pOdd1 Loop vector based on pSB3K3. | ||
| − | a | + | New pOdd backbones for Level 3 assemblies were constructed using the Gibson assembly method. These are three sets of pOdd plasmids which suit different situations: |
| + | <html> | ||
| + | <ul> | ||
| + | <li>pSB3K0# (</html>[[Part:BBa_K3425005|<partinfo>BBa_K3425005</partinfo>]]<html> to </html>[[Part:BBa_K3425008|<partinfo>BBa_K3425008</partinfo>]]<html>): medium copy number (10-12 copies)</li> | ||
| + | <li>pSB4K0# (</html>[[Part:BBa_K3425009|<partinfo>BBa_K3425009</partinfo>]]<html> to </html>[[Part:BBa_K3425012|<partinfo>BBa_K3425012</partinfo>]]<html>): low copy number (~5 copies) </li> | ||
| + | <li>pSB4A0# (</html>[[Part:BBa_K3425013|<partinfo>BBa_K3425013</partinfo>]]<html> to </html>[[Part:BBa_K3425016|<partinfo>BBa_K3425016</partinfo>]]<html>): low copy number (~5 copies) with different resistance for cotransformations</li> | ||
| + | </ul> | ||
| + | </html> | ||
| + | |||
| + | ===Medium copy number pOdd Loop Vectors (for Level 3)=== | ||
| + | {{CSS/DNA_Table}} | ||
| + | |||
| + | When Level 2 multi-transcriptional units (MTUs) are not enough to hold your whole system, you might want to construct higher level assemblies. The current pOdd backbones, pSB1K0#, are high copy number backbones (100-300 copies) which are not suitable for Level 3 assemblies. The metabolic burden caused by the expression of multiple transcriptional units in a high copy number plasmid can be too overwhelming for the cell, which might make the plasmid unstable and prone to mutation or loss [1]. For this reason, new sets of medium (10-12 copies) and low (~5 copies) copy number pOdd backbones were designed and assembled. | ||
| + | |||
| + | This page details the set of medium copy number backbones. The pSB3K01-pSB3K04 series of plasmids is based on the pOdd set of iGEM Type IIS Vectors and are built upon the pSB3K3 plasmid backbone. When four Level 2 MTUs are assembled into these plasmid backbones (with <partinfo>BBa_J04454</partinfo>), the multi-MTU will be flanked by SapI restriction sites and the same 3 bp Fusion Sites as Level 1 assemblies. | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <<table class="dnaTable"> | ||
| + | <tr> | ||
| + | <th>Part number</th> | ||
| + | <th>Name</th> | ||
| + | <th>Loop Name</th> | ||
| + | <th>Fusion Site 5'</th> | ||
| + | <th>Multi-MTU</th> | ||
| + | <th>Fusion Site 3'</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></html>[[Part:BBa_K3425005|<partinfo>BBa_K3425005</partinfo>]]<html></td> | ||
| + | <td>pSB3K01</td> | ||
| + | <td>pOdd1</td> | ||
| + | <td class="dnaSeq rightAlign">ATG</td> | ||
| + | <td class="centerAlign partType" style="background-color: #C00001;">Multi-MTU 1</td> | ||
| + | <td class="dnaSeq">GCA</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></html>[[Part:BBa_K3425006|<partinfo>BBa_K3425006</partinfo>]]<html></td> | ||
| + | <td>pSB3K02</td> | ||
| + | <td>pOdd2</td> | ||
| + | <td class="dnaSeq rightAlign">GCA</td> | ||
| + | <td class="centerAlign partType" style="background-color: #44546A;">Multi-MTU 2</td> | ||
| + | <td class="dnaSeq">TAC</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></html>[[Part:BBa_K3425007|<partinfo>BBa_K3425007</partinfo>]]<html>/td> | ||
| + | <td>pSB3K03</td> | ||
| + | <td>pOdd3</td> | ||
| + | <td class="dnaSeq rightAlign">TAC</td> | ||
| + | <td class="centerAlign partType" style="background-color: #FFC001;">Multi-MTU 3</td> | ||
| + | <td class="dnaSeq">CAG</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></html>[[Part:BBa_K3425008|<partinfo>BBa_K3425008</partinfo>]]<html></td> | ||
| + | <td>pSB3K04</td> | ||
| + | <td>pOdd4</td> | ||
| + | <td class="dnaSeq rightAlign">CAG</td> | ||
| + | <td class="centerAlign partType" style="background-color: #7030A0;">Multi-MTU 4</td> | ||
| + | <td class="dnaSeq">GGT</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </table> | ||
| + | </center> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | {{TypeIIS/References}} | ||
| + | |||
| + | -'''Note:''' This section was partially adapted from [[Part:pSB1K01|<partinfo>pSB1K01</partinfo>]]. For more information on Level 1 assemblies, you can visit [[Part:pSB1K01|<partinfo>pSB1K01</partinfo>]]. For more information as well as all our experience in this cloning method you can visit Team Uppsala 2020's <html><a href="https://2020.igem.org/Team:UofUppsala/Engineering#TypeIIS">iGEM Type IIS standard guidebook</a></html>. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 9: | Line 77: | ||
<!-- --> | <!-- --> | ||
| − | <span class='h3bb'>Sequence and Features</span> | + | <html><span class='h3bb'><b>Sequence and Features</b></span></html> |
<partinfo>BBa_K3425005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3425005 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | ===Gibson assembly of pOdd Loop vectors=== | ||
| + | |||
| + | <html> | ||
| + | |||
| + | <p>Each set of new pOdd backbones comprises four plasmids which have the mRFP1 device for pOdd plasmids (</html><partinfo>BBa_J04454</partinfo><html>) flanked by the corresponding Type IIS fusion sites substituting the BioBrick sites of the base plasmid. This set, pSB3K0#, is derived from the </html><partinfo>pSB3K3</partinfo><html> plasmid and has the same copy number as the existing pEven backbones (10-12 copies).</p> | ||
| + | |||
| + | <p>Gibson assembly was used to construct these backbones, and the details can be found in the Design page of each of these parts. Afterwards, it was needed to screen for plasmids which contained the mRFP1 device and had the correct antibiotic resistance and copy number. Antibiotic resistance was screened by plating the transformation, the presence of the mRFP1 device by PCR and the copy number by sequencing the ori and by controlled plasmid extraction.</p> | ||
| + | |||
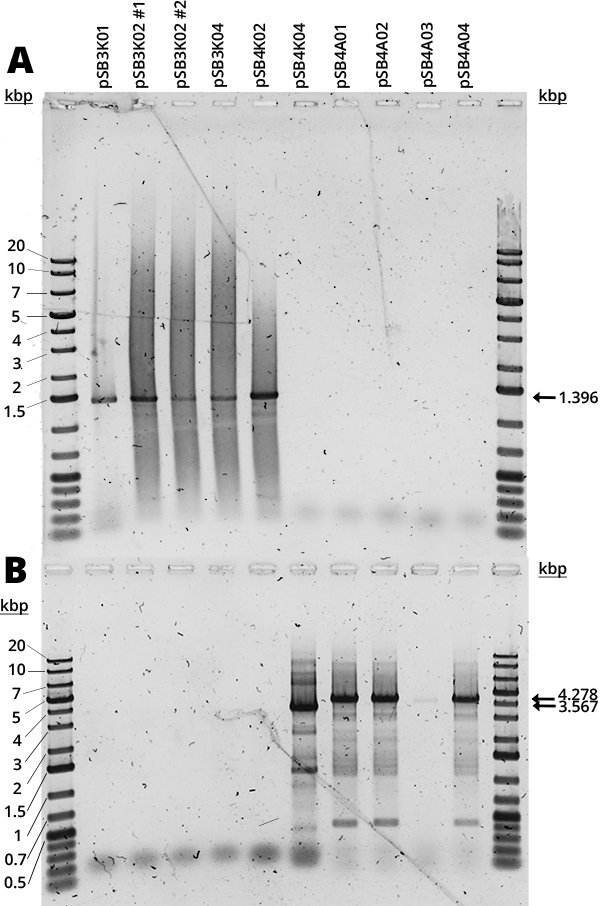
| + | <p>In Figure 1 can be seen the amplification of the RFP device (A) and the full plasmid (B). The mRFP1 device (1396bp amplification) was present in all available pSB3K0# and pSB4K02. The full backbone amplification (3756bp for pSB3K0#, 3567bp for pSB4K0# and 4278bp for pSB4A0#) is positive for pSB4K02 and all pSB4A0#. Most samples (Figure 1, B) have multiple bands which we believe are non-specific and are due to the long extension times required for full backbone amplification, since the strongest band corresponds to the expected size.</p> | ||
| + | |||
| + | <p>The smear that can be seen in all samples we suspect is due to the use of Diamond Dye (Promega), as it is something we have consistently observed with this DNA dye. Furthermore, it is concerning how in each gel half of the samples were negative, but it was different samples each time. Judging by the other results (present in Team Uppsala 2020's <a href="https://2020.igem.org/Team:UofUppsala/Results#TypeIIS">Wet lab</a> page), it seems to be an error in the handling of samples. It would be good to repeat it, but there was no time. From the available results, however, it seems that all backbones are correct.</p> | ||
| + | |||
| + | <figure> | ||
| + | <p><img src="https://2020.igem.org/wiki/images/d/d3/T--UofUppsala--20201001_gibson.png" width="100%" height="" alt=""> | ||
| + | <figcaption>Figure 2. 1% Agarose Gel Electrophoresis - Diamond Nucleic Acid Dye analysis of (A) VF2-VR PCR and (B) full plasmid. (1, 12) MW marker (GeneRuler 1kb Plus DNA Ladder), (2) pSB3K01, (3) pSB3K02 colony #1, (4) pSB3K02 colony #2, (5) pSB3K04, (6) pSB4K02, (7) pSB4K04, (8) pSB4A01, (9) pSB4A02, (10) pSB4A03, (11) pSB4A04.</figcaption> | ||
| + | </figure> | ||
| + | |||
| + | <p>For further information on this experiment along with the sequencing results and copy number results, please visit Team Uppsala 2020's <a href="https://2020.igem.org/Team:UofUppsala/Results#TypeIIS">Wet lab</a> page.</p> | ||
| + | </html> | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
| Line 17: | Line 105: | ||
<partinfo>BBa_K3425005 parameters</partinfo> | <partinfo>BBa_K3425005 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ===References=== | ||
| + | [1] Jones, K. L., Kim, S.-W., and Keasling, J. D. (2000) Low-Copy Plasmids can Perform as Well as or Better Than High-Copy Plasmids for Metabolic Engineering of Bacteria. Metabolic Engineering. 2, 328–338 | ||
Revision as of 09:24, 21 October 2020
pSB3K01: pOdd1 Loop Vector based on pSB3K3
This, pSB3K01, part is a medium copy number pOdd1 Loop vector based on pSB3K3.
New pOdd backbones for Level 3 assemblies were constructed using the Gibson assembly method. These are three sets of pOdd plasmids which suit different situations:
- pSB3K0# (BBa_K3425005 to BBa_K3425008): medium copy number (10-12 copies)
- pSB4K0# (BBa_K3425009 to BBa_K3425012): low copy number (~5 copies)
- pSB4A0# (BBa_K3425013 to BBa_K3425016): low copy number (~5 copies) with different resistance for cotransformations
Medium copy number pOdd Loop Vectors (for Level 3)
When Level 2 multi-transcriptional units (MTUs) are not enough to hold your whole system, you might want to construct higher level assemblies. The current pOdd backbones, pSB1K0#, are high copy number backbones (100-300 copies) which are not suitable for Level 3 assemblies. The metabolic burden caused by the expression of multiple transcriptional units in a high copy number plasmid can be too overwhelming for the cell, which might make the plasmid unstable and prone to mutation or loss [1]. For this reason, new sets of medium (10-12 copies) and low (~5 copies) copy number pOdd backbones were designed and assembled.
This page details the set of medium copy number backbones. The pSB3K01-pSB3K04 series of plasmids is based on the pOdd set of iGEM Type IIS Vectors and are built upon the pSB3K3 plasmid backbone. When four Level 2 MTUs are assembled into these plasmid backbones (with BBa_J04454), the multi-MTU will be flanked by SapI restriction sites and the same 3 bp Fusion Sites as Level 1 assemblies.
| Part number | Name | Loop Name | Fusion Site 5' | Multi-MTU | Fusion Site 3' |
|---|---|---|---|---|---|
| BBa_K3425005 | pSB3K01 | pOdd1 | ATG | Multi-MTU 1 | GCA |
| BBa_K3425006 | pSB3K02 | pOdd2 | GCA | Multi-MTU 2 | TAC |
| BBa_K3425007/td> | pSB3K03 | pOdd3 | TAC | Multi-MTU 3 | CAG |
| BBa_K3425008 | pSB3K04 | pOdd4 | CAG | Multi-MTU 4 | GGT |
The iGEM Type IIS assembly standard is based on MoClo and Loop
- A Modular Cloning System for Standardized Assembly of Multigene Constructs
- Loop assembly: a simple and open system for recursive fabrication of DNA circuits
-Note: This section was partially adapted from pSB1K01. For more information on Level 1 assemblies, you can visit pSB1K01. For more information as well as all our experience in this cloning method you can visit Team Uppsala 2020's iGEM Type IIS standard guidebook.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2723
Illegal PstI site found at 12 - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2723
Illegal NheI site found at 1378
Illegal PstI site found at 12 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2723
Illegal XhoI site found at 171
Illegal XhoI site found at 1014 - 23INCOMPATIBLE WITH RFC[23]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2723
Illegal PstI site found at 12 - 25INCOMPATIBLE WITH RFC[25]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2723
Illegal PstI site found at 12
Illegal AgeI site found at 1464
Illegal AgeI site found at 1787 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal SapI site found at 2729
Illegal SapI.rc site found at 5
Gibson assembly of pOdd Loop vectors
Each set of new pOdd backbones comprises four plasmids which have the mRFP1 device for pOdd plasmids (BBa_J04454) flanked by the corresponding Type IIS fusion sites substituting the BioBrick sites of the base plasmid. This set, pSB3K0#, is derived from the pSB3K3 plasmid and has the same copy number as the existing pEven backbones (10-12 copies).
Gibson assembly was used to construct these backbones, and the details can be found in the Design page of each of these parts. Afterwards, it was needed to screen for plasmids which contained the mRFP1 device and had the correct antibiotic resistance and copy number. Antibiotic resistance was screened by plating the transformation, the presence of the mRFP1 device by PCR and the copy number by sequencing the ori and by controlled plasmid extraction.
In Figure 1 can be seen the amplification of the RFP device (A) and the full plasmid (B). The mRFP1 device (1396bp amplification) was present in all available pSB3K0# and pSB4K02. The full backbone amplification (3756bp for pSB3K0#, 3567bp for pSB4K0# and 4278bp for pSB4A0#) is positive for pSB4K02 and all pSB4A0#. Most samples (Figure 1, B) have multiple bands which we believe are non-specific and are due to the long extension times required for full backbone amplification, since the strongest band corresponds to the expected size.
The smear that can be seen in all samples we suspect is due to the use of Diamond Dye (Promega), as it is something we have consistently observed with this DNA dye. Furthermore, it is concerning how in each gel half of the samples were negative, but it was different samples each time. Judging by the other results (present in Team Uppsala 2020's Wet lab page), it seems to be an error in the handling of samples. It would be good to repeat it, but there was no time. From the available results, however, it seems that all backbones are correct.
For further information on this experiment along with the sequencing results and copy number results, please visit Team Uppsala 2020's Wet lab page.
References
[1] Jones, K. L., Kim, S.-W., and Keasling, J. D. (2000) Low-Copy Plasmids can Perform as Well as or Better Than High-Copy Plasmids for Metabolic Engineering of Bacteria. Metabolic Engineering. 2, 328–338
