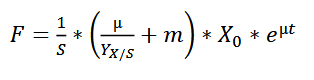Difference between revisions of "Part:BBa K3264005"
(→Usage and Biology) |
|||
| Line 30: | Line 30: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K3264005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3264005 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | == iGEM 2022 UCopenhagen - Contribution == | ||
| + | |||
| + | '''Designers:''' Akila Ravikumar and Matteo Soana | ||
| + | |||
| + | '''Team:''' Netlantis | ||
| + | |||
| + | === Large scale production of NT-4rep-CT === | ||
| + | |||
| + | As part of our project, we produced two minispidroins whose production was scaled up. One of these minispidroins had the same amino acid sequence as this part (we used the same scale up strategy for all of them). Herein, we provide information regarding the fermentation of this protein using E.coli in a fed-batch cultivation. Our entire fermentation process was based on the methods of the paper: “High-yield production of a super-soluble miniature spidroin for biomimetic high-performance materials” by Schmuck et al., 2021. | ||
| + | |||
| + | '''Composition of Batch and Fed-batch media''' | ||
| + | |||
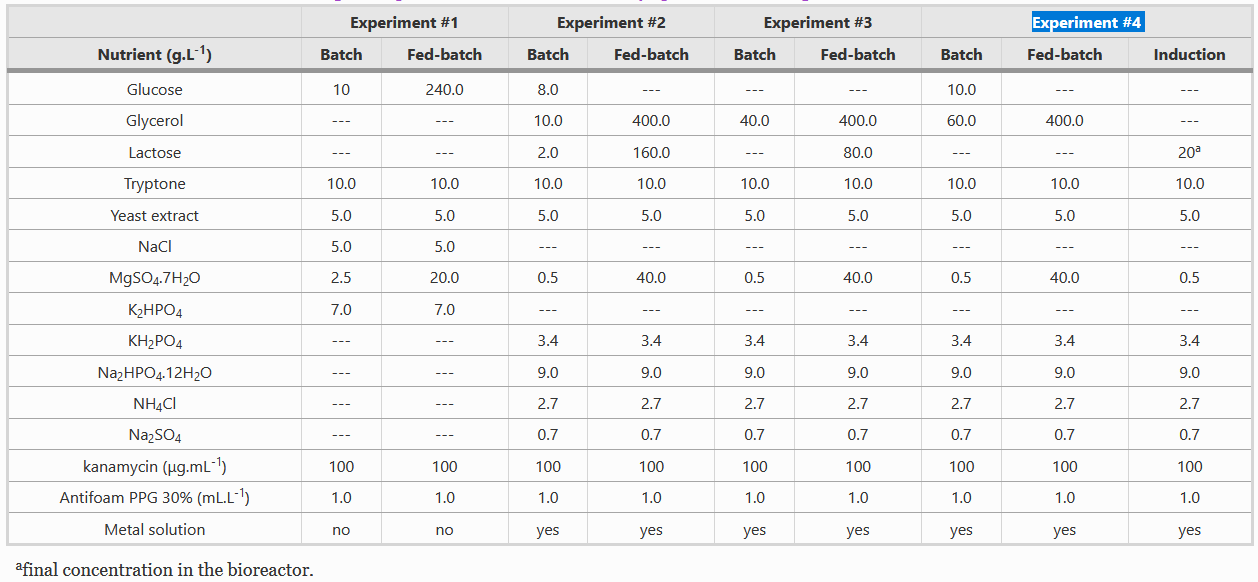
| + | Instead of LB media, we used a complex growth medium that integrated a specific trace metal solution. The authors obtained this media from experiment #4 (see below) of another paper: A.J. da Silva, A.C.L. Horta, A.M. Velez, M.R.C. Iemma, C.R. Sargo, R.L.C. Giordano, M.T.M. Novo, R.C. Giordano, T.C. Zangirolami, Springerplus 2 (2013) 1–12. | ||
| + | |||
| + | [[File:bronze1.png|900px]] | ||
| + | |||
| + | For our bioreactor, we always started cultures with 1 L of batch media and set the system such that 500 mL of fed-batch media would enter the system at a specific rate and we used Silica antifoam. | ||
| + | |||
| + | The feeding rate should follow the formula: | ||
| + | |||
| + | [[File:bronze2.png]] | ||
| + | |||
| + | wherein, | ||
| + | |||
| + | F - the rate of feeding (l h-1 ) | ||
| + | |||
| + | S - concentration of the glycerol in the feed (g l-1 ) | ||
| + | |||
| + | µ - specific growth rate (h-1) | ||
| + | |||
| + | YX/S - biomass yield on the substrate (g g-1 ) | ||
| + | |||
| + | m - specific maintenance coefficient (g g-1 h -1 ) | ||
| + | |||
| + | X - biomass concentration (g l-1 ) | ||
| + | |||
| + | For the fermentation of this minispidroin protein, the authors set µ to 0.1, m to 0.025, and YX/S to 0.622. | ||
| + | |||
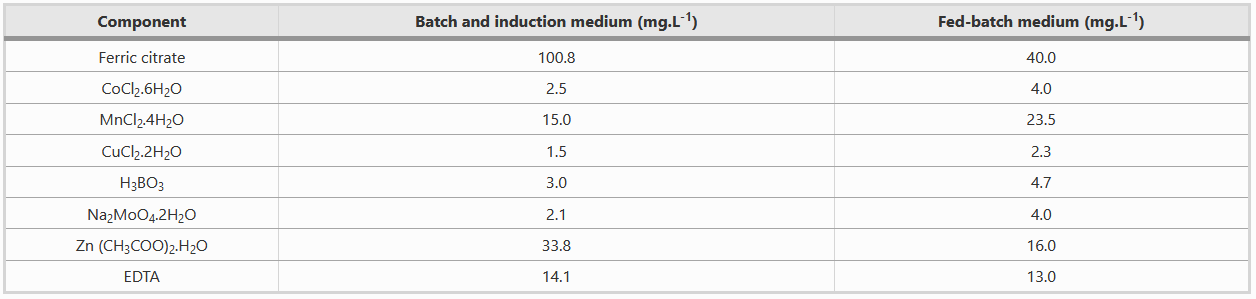
| + | '''Composition of Metal solution''' | ||
| + | |||
| + | Both batch and fed-batch media have to be complemented with two different metal solutions, as shown in the table below. | ||
| + | |||
| + | [[File:bronze3.png|900px]] | ||
| + | |||
| + | ''Note:'' Be careful with the preparation of ferric citrate because residual non solubilised Fe(3+), being rust, can be quite problematic for the small fermentor tubings and clog them, which results in the interruption of the release of fed-batch media. | ||
| + | |||
| + | '''Autoclaving''' | ||
| + | |||
| + | Before fermentation, it is important to autoclave all media and filter all solutions. It is important to not autoclave some solutions since they have heat labile compounds and hence, they can be added to the autoclaved media later. We filtered, rather than autoclave, the glucose (700 g/l), the trace metal solution (25x), and MgSO4 (245 g/l). Kanamycin was also added later. Further, unlike the paper where they add the antifoam to the media, we used the antifoam sensor of the fermenter to add it when necessary at the default flow rate. | ||
| + | |||
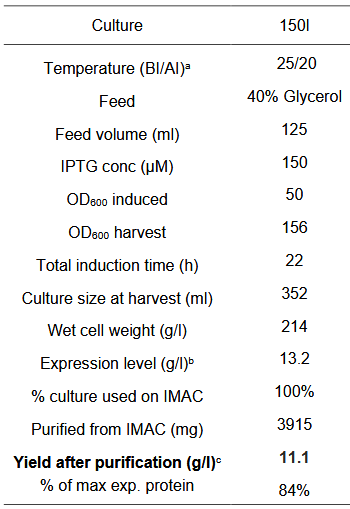
| + | '''Parameters for the Fermentation cycle''' | ||
| + | |||
| + | Our fermentation strategy worked in every protein production cycle, but we had to slightly modify the original one (below). | ||
| + | |||
| + | [[File:bronze4.png]] | ||
| + | |||
| + | Since glucose inhibits protein expression of T7 expression vectors, the full consumption of glucose has to occur before the beginning of protein expression. Full oxygen consumption is visible as a spike in the saturated oxygen levels (pO2) in the bioreactor trends. In our 3L bioreactor, this tends to happen around 20h and at this point, both induction (150 uM IPTG) and drop in temperature (29->20) should be done. The protein production was stopped after about 20h with a total of 40h of fermentation time per production cycle. | ||
| + | |||
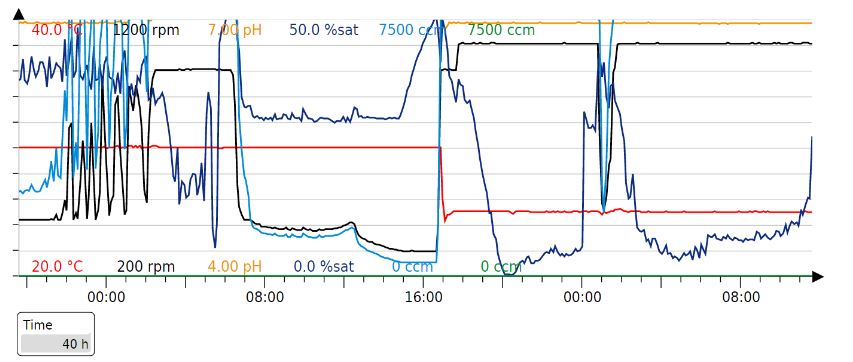
| + | Our production cycle (40h): | ||
| + | |||
| + | [[File:Ferm 2r.jpeg]] | ||
| + | |||
| + | For more information, please refer to part <partinfo>BBa_K4247012</partinfo> (Minispidroin_NT-4rep-CT_N-6His) where the various characterizations we performed on this protein can be found in detail. | ||
Revision as of 10:59, 7 October 2022
NT4RepCT
NT-4Rep-CT is the recombinant spider silk protein (spidroin) that can spin into artificial spider silk fibers. This part is in the part collection where we provide improved recombinant chromoproteins and spidroins based on the functions of different regions of naturally occurring spidroin. By carrying out different mixing of the proteins of different combinations, we can produce artificially spun silk of different colors and properties.
The part collection includes: Parts that are different kinds of spidroins for silk spining. BBa_K3264000 BBa_K3264005 BBa_K3264007 BBa_K3264009 BBa_K3264018. BBa_K3264019. Parts that for combining with the spidroins to form color silks BBa_K3264012. BBa_K3264013. BBa_K3264014. BBa_K3264015. BBa_K3264016. BBa_K3264017. Part for adding more extensive repetitive region to the spidroin protein BBa_K3264011.
Our part collection can help and inspire future teams to further design recombinant spider silks of more variety and give silks diversified color and property to fulfill the collection.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 943
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 424
Illegal SpeI site found at 943 - 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 943
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 943
- 1000COMPATIBLE WITH RFC[1000]
iGEM 2022 UCopenhagen - Contribution
Designers: Akila Ravikumar and Matteo Soana
Team: Netlantis
Large scale production of NT-4rep-CT
As part of our project, we produced two minispidroins whose production was scaled up. One of these minispidroins had the same amino acid sequence as this part (we used the same scale up strategy for all of them). Herein, we provide information regarding the fermentation of this protein using E.coli in a fed-batch cultivation. Our entire fermentation process was based on the methods of the paper: “High-yield production of a super-soluble miniature spidroin for biomimetic high-performance materials” by Schmuck et al., 2021.
Composition of Batch and Fed-batch media
Instead of LB media, we used a complex growth medium that integrated a specific trace metal solution. The authors obtained this media from experiment #4 (see below) of another paper: A.J. da Silva, A.C.L. Horta, A.M. Velez, M.R.C. Iemma, C.R. Sargo, R.L.C. Giordano, M.T.M. Novo, R.C. Giordano, T.C. Zangirolami, Springerplus 2 (2013) 1–12.
For our bioreactor, we always started cultures with 1 L of batch media and set the system such that 500 mL of fed-batch media would enter the system at a specific rate and we used Silica antifoam.
The feeding rate should follow the formula:
wherein,
F - the rate of feeding (l h-1 )
S - concentration of the glycerol in the feed (g l-1 )
µ - specific growth rate (h-1)
YX/S - biomass yield on the substrate (g g-1 )
m - specific maintenance coefficient (g g-1 h -1 )
X - biomass concentration (g l-1 )
For the fermentation of this minispidroin protein, the authors set µ to 0.1, m to 0.025, and YX/S to 0.622.
Composition of Metal solution
Both batch and fed-batch media have to be complemented with two different metal solutions, as shown in the table below.
Note: Be careful with the preparation of ferric citrate because residual non solubilised Fe(3+), being rust, can be quite problematic for the small fermentor tubings and clog them, which results in the interruption of the release of fed-batch media.
Autoclaving
Before fermentation, it is important to autoclave all media and filter all solutions. It is important to not autoclave some solutions since they have heat labile compounds and hence, they can be added to the autoclaved media later. We filtered, rather than autoclave, the glucose (700 g/l), the trace metal solution (25x), and MgSO4 (245 g/l). Kanamycin was also added later. Further, unlike the paper where they add the antifoam to the media, we used the antifoam sensor of the fermenter to add it when necessary at the default flow rate.
Parameters for the Fermentation cycle
Our fermentation strategy worked in every protein production cycle, but we had to slightly modify the original one (below).
Since glucose inhibits protein expression of T7 expression vectors, the full consumption of glucose has to occur before the beginning of protein expression. Full oxygen consumption is visible as a spike in the saturated oxygen levels (pO2) in the bioreactor trends. In our 3L bioreactor, this tends to happen around 20h and at this point, both induction (150 uM IPTG) and drop in temperature (29->20) should be done. The protein production was stopped after about 20h with a total of 40h of fermentation time per production cycle.
Our production cycle (40h):
For more information, please refer to part BBa_K4247012 (Minispidroin_NT-4rep-CT_N-6His) where the various characterizations we performed on this protein can be found in detail.
Usage and Biology
Spider silk serves as a new material with superior properties that can be applied in medication, cloth, and aerospace fields. However, spider breeding is not applicable due to the spider's fierce behavior. The current approach is to produce recombinant spidroins (silk proteins) from other chassis and spin them into silk.
Enable a step closer to unobtainable size (556 kDa) of natural spider spidroin, containing 192 repeat motifs of the Nephila clavipes dragline spidroin, we have developed a synthetic biology approach where we combine standardized DNA part assembly and split the intermediate ligation to produce recombinant spidroins of NT-4Rep-CT, in this case, extra a 2Rep is added to the NT-2Rep-CT and was expected to have a better mechanical performance after spinning. NT-2Rep-CT is the common architecture in two main types of spider silk proteins: major ambulate spidorins(MaSps) and minor ampullate spidorins (MiSps). NT is a non-repetitive N-terminal domain, as well as CT, the C-terminal domain. In between them is an extensive repetitive region-Rep In the storage of spider silk protein, NT is a monomeric and highly soluble region in neutral pH, and it provides the solubility of the entire spidroin. NT can form stable dimers when pH decreases in the spinning duct. On the other side during the decrease of pH, the CT gets disrupted and unfold to turn in the beta-sheet amyloid-like fibrils. It is hypothesized that structural change of CT precipitates the change of the repetitive region to beta-sheet conformation.
Reference
[1]Andersson, Marlene, et al. “Biomimetic Spinning of Artificial Spider Silk from a Chimeric Minispidroin.” /Nature Chemical Biology/, vol. 13, no. 3, Sept. 2017, pp. 262–264., doi:10.1038/nchembio.2269.
[2]Zhou, Yizhong, et al. “Production and Properties of Triple Chimeric Spidroins.” Biomacromolecules, vol. 19, no. 7, 2018, pp. 2825–2833., doi:10.1021/acs.biomac.8b00402.
Characterization
We aimed to use a synthetic biology approach to combine standardized DNA part NT-4Rep-CT, and transform constructs to metabolically engineered E. coli for bioproduction.
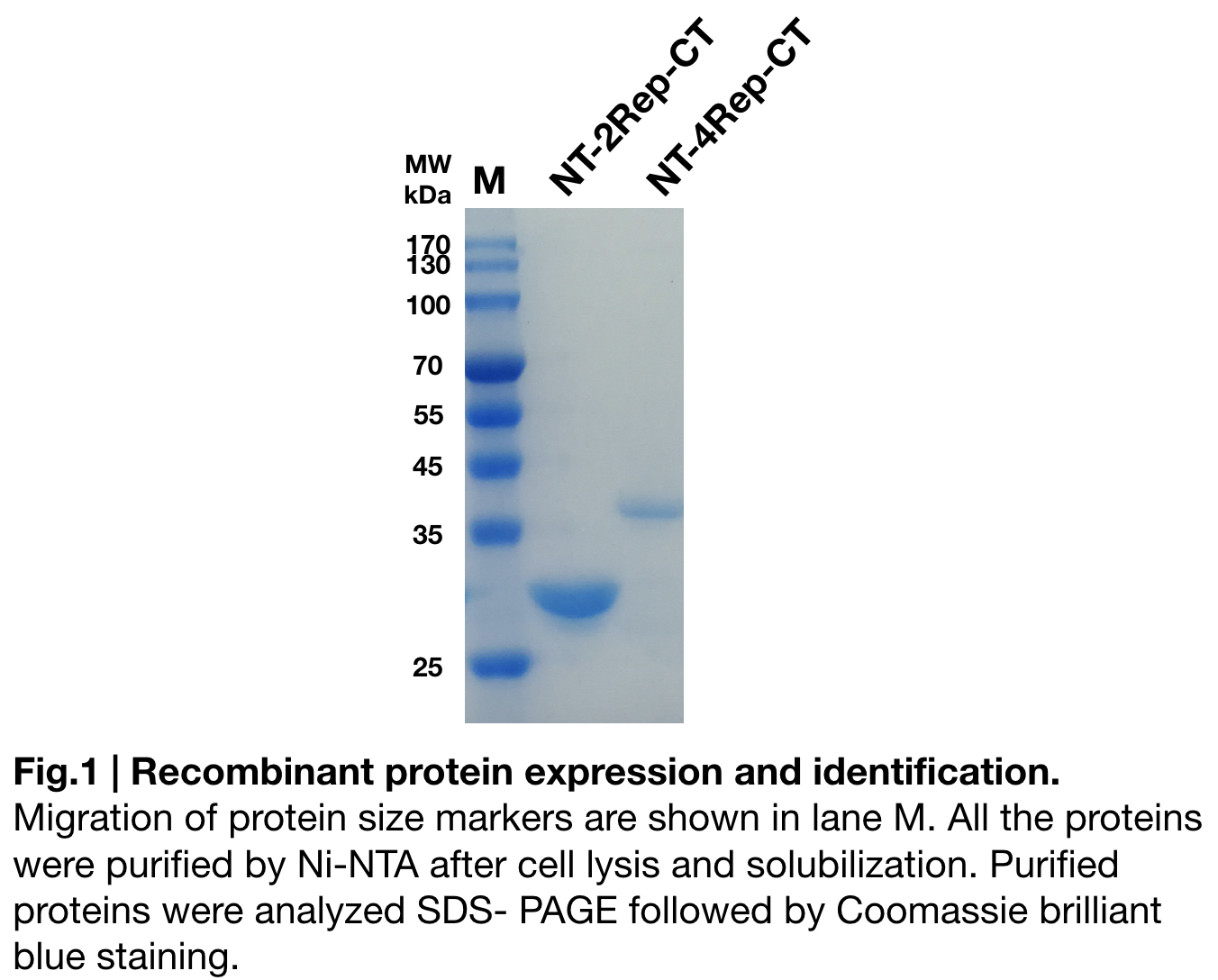
Purification and SDS PAGE
In order to detect whether NT-4Rep-CT protein expression was induced by adding isopropylthiogalactoside (final concentration 0.3mM), we used SDS-page(12%) to determine the presence of target protein. Target protein, NT-4Rep-CT was successfully induced and resulted in a higher molecular weight as it shown on the SDS-page than NT-2Rep-CT (Fig.1). The increase in molecular weight is regarded as reasonable because of the 2Rep added to the original NT-2Rep-CT.