|
|
| Line 44: |
Line 44: |
| | | | |
| | <html> | | <html> |
| − | </style> | + | <h4>Cyanobacterial shuttle vectors </h4> |
| − | <div>
| + | |
| − | <div class="box-dark">
| + | As we have already clarified in the description part, self replicating shuttle vectors are essential for many |
| − | <hr class="line">
| + | workflows, as the gene expression levels are higher and non of the tedious selection processes that come with |
| − |
| + | genomic integrations have to be done. <br> |
| − | </div>
| + | |
| − | <div style="margin-top: 10vh;">
| + | On our road to the modular vector we were seeking, we firstly cured our own S. elongatus UTEX 2973 strain of its |
| − | <section class="section">
| + | pANS plasmid. This was done by transforming the pAM4787 vector, which holds a spectinomycin resistance as well as a |
| − | <h1 class="title">The origin</h1>
| + | YFP cassette |
| − | <p style="text-align: justify;">
| + | <a href= https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.000377> (Chen et al., 2016)</a>. |
| − | Inspired by the fast progress in Synthetic Biology and its urgent need for genetic tools that enable the
| + | Due to plasmid incompatibility - explained here in our design section <b>[Link to shuttle vector design]</b> - and because antibiotic pressure is applied, the pANS plasmid was over time cured from the |
| − | exploitation of cyanobacteria for research and biotechnological applications, we set out to construct the most
| + | strain, which then just kept the pAM4787 plasmid. Transformation was done by conjugation with the pRK2013 plasmid in |
| − | versatile shuttle vector for cyanobacteria based on the modular Golden Gate Assembly method, allowing for flexible
| + | DH5ɑ and the pAM4787 in HB101. Both were grown to an OD600≈0.5, washed in LB and mixed with S. elongatus which was |
| − | cloning into a reliable self-replicating system.<br>
| + | grown to late exponential phase and then washed in BG11. |
| − | <br>
| + | We could clearly show, that the conjugant strain bears the pAM4787 plasmid if selective pressure is held up. |
| − | </p>
| + | <figure Style="text-align:center"> |
| − | <figure>
| + | <img style="height: 65ex; width: 50ex" |
| − | <img style="display: block; margin: 0 auto 0 auto; width:50%"
| + | src= https://2019.igem.org/wiki/images/e/eb/T--Marburg--YFPconstructConjugantFACS.png |
| − | src="https://lh3.googleusercontent.com/8ko4suiu3NQ_qmRIeZf1k1sg5EUw8g4JXfkGG3xAmRk1dxaVlZQbzC9Uz-6ToGKXaAf5p_yx9MVHhlO3QdMmG_l0ukJ0OVQOWBzcouM-HOTc_ta7LblxiVtTdLKrf9q4bpzP6ZRP"
| + | alt=" https://2019.igem.org/wiki/images/e/eb/T--Marburg--YFPconstructConjugantFACS.png"> |
| − | alt="Lvl1 ori">
| + | <figcaption> |
| − | <figcaption>
| + | Fig. 11: Cell counts of conjugant strain. The y axis shows relative YFP fluorescence and the x axis relative autofluorescence. |
| − | Fig.1 - Lvl1 ori
| + | </figcaption> |
| − | </figcaption>
| + | </figure> |
| − | </figure>
| + | This was followed by us starting to culture the pAM4787 bearing strain without |
| − |
| + | antibiotics again, slowly removing selective pressure from the cells. As the plasmid does not give them any other |
| − | <div><p>
| + | advantage and is probably just more metabolic burden due to the constantly produced YFP proteins it is slowly being |
| − | Introduction of exogenous DNA can be done in multiple ways and propagated in a strain if it is integrated in the | + | lost. |
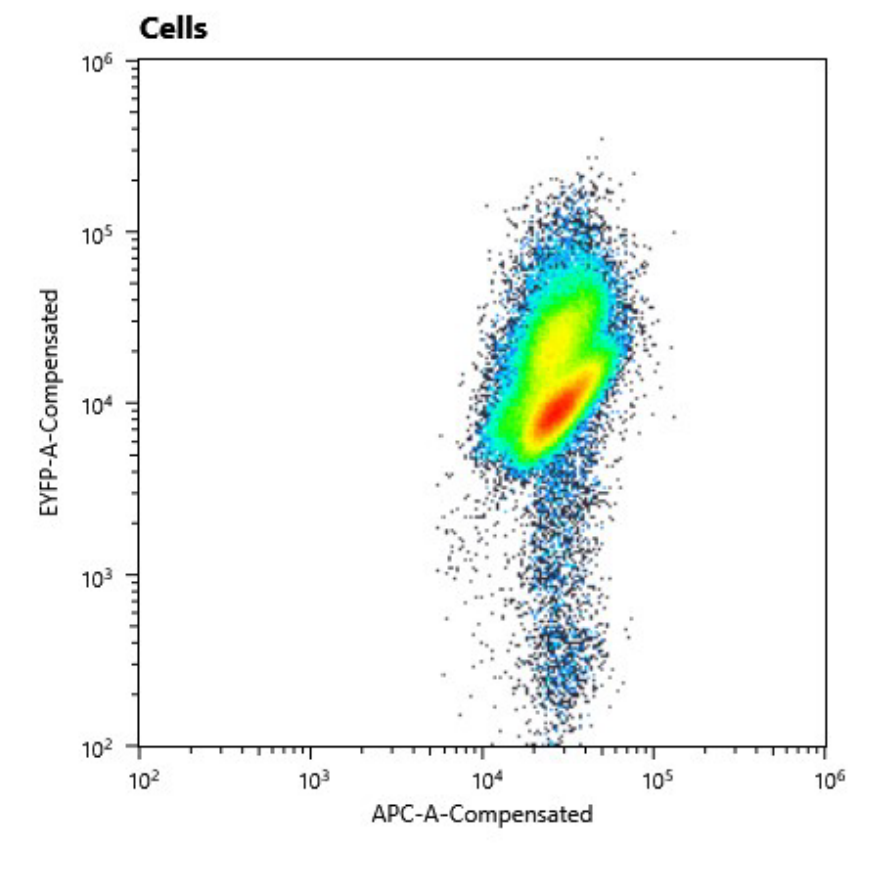
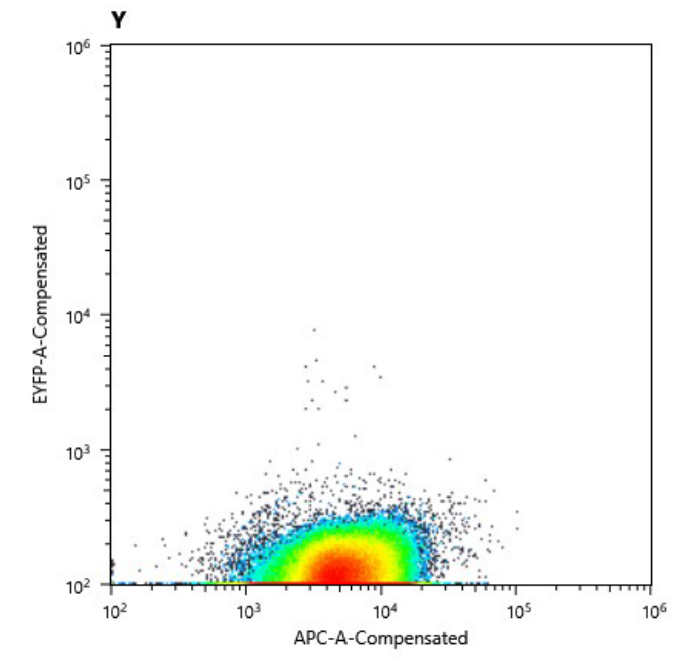
| − | chromosome or stably expressed on a self-replicating plasmid.<br> For rapid prototyping in cyanobacteria | + | We could prove this in multiple setups: with the flow cytometry device we were kindly granted access to we could |
| − | self-replicating plasmids are of higher interest than genome-integrations, as the latter can be quite | + | clearly show the missing YFP signal in the cured <i>S. elongatus </i>strain and |
| − | time-consuming in cyanobacterial strains with multiple genome copies (<a | + | logically this could also be observed over our UV table |
| − | href="https://www.ncbi.nlm.nih.gov/pubmed/22092711">Griese <i>et al.,</i> 2011</a>). Furthermore, genes
| + | |
| − | introduced in self-replicating vectors have been shown to have higher gene-expression levels than those integrated
| + | <figure Style="text-align:center"> |
| − | in the genome, as copy numbers are typically higher (<a href="https://doi.org/10.1099/mic.0.000377">Chen Titel anhand dieser DOI in Citavi-Projekt übernehmen <i>et
| + | <img style="height: 65ex; width: 50ex" |
| − | al.,</i> 2016</a>) – a desirable trait, not just for rapid prototyping in research applications, but also for
| + | src= https://2019.igem.org/wiki/images/c/c4/T--Marburg--CuredStrainsFACS.png |
| − | biotechnological production of valuable compounds.<br>
| + | alt="https://2019.igem.org/wiki/images/c/c4/T--Marburg--CuredStrainsFACS.png "> |
| − | With our shuttle-vectors encompass a cyanobacterial origin of replication (ori) from <i>Synechococcus
| + | <img style="height: 65ex; width: 50ex" |
| − | elongatus</i> PCC7942 as well as an <i>E.coli</i> ori, which is perfect for fast cloning processes, as these
| + | src= https://2019.igem.org/wiki/images/d/da/T--Marburg--CuredStrainUV.png |
| − | vectors can be easily recovered from the cyanobacteria and reintroduced in an <i>E.coli</i> strain.<br> | + | alt="https://2019.igem.org/wiki/images/d/da/T--Marburg--CuredStrainUV.png"> |
| − | <br>
| + | <figcaption> |
| − | </p>
| + | a) Cell counts of cured strain. The y axis shows relative YFP fluorescence and the x axis relative autofluorescence |
| − | <br>
| + | b) Comparison of the fluorescence signal of the transformed (left) and cured (right) strain. |
| − | <p style="font-size: 20px">
| + | </figcaption> </figure> |
| − | Currently existing shuttle vectors for cyanobacteria are still based on standard systems working with multiple
| + | |
| − | cloning sites (MCS) for expression of homologous genes (<a href="https://doi.org/10.1099/mic.0.000377">Chen Titel anhand dieser DOI in Citavi-Projekt übernehmen <i>et
| + | |
| − | al.,</i> 2016</a>). A huge downside is that these vectors include either an MCS (e.g. pAM5188) or a
| + | |
| − | fluorescence reporter (e.g. pAM4787), which is unpractical for easy selection of recombinant clones. Additionally,
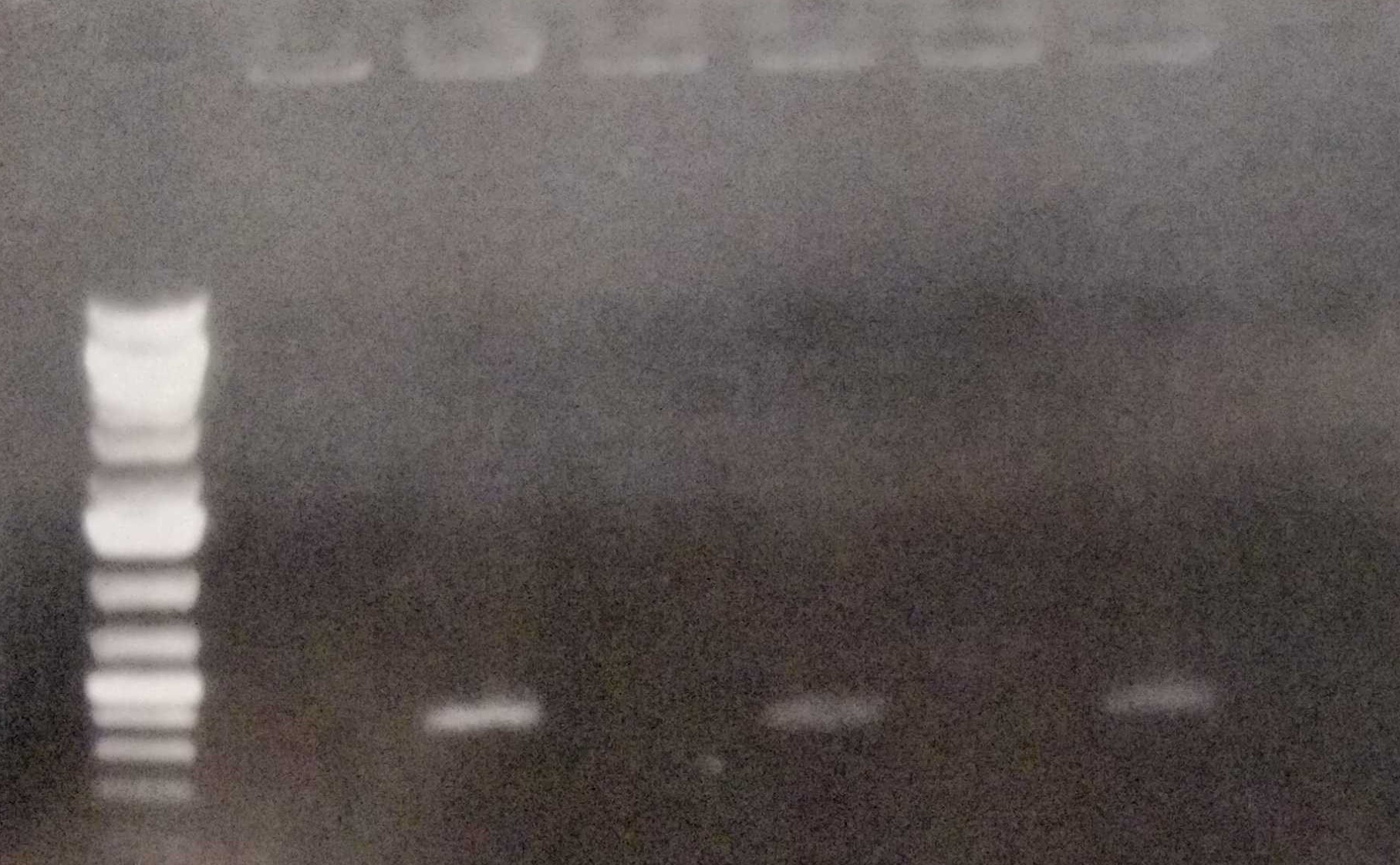
| + | Furthermore we performed colony PCRs as a test. We sent our plasmid-free strain to Next Generation Sequencing in order to ensure that the strain really has lost the |
| − | an MCS comes with possible sequence constraints due to restriction sites leaving unwanted base pairs in your
| + | pANS plasmid. |
| − | constructs.<br>
| + | <figure Style="text-align:center"> |
| − | Facilitating and standardizing the process of engineering biological systems is one of the fundamental goals of
| + | <img style="height: 65ex; width: 50ex" |
| − | synthetic biology (<a href="https://doi.org/10.1186/1754-1611-2-5">Shetty Titel anhand dieser DOI in Citavi-Projekt übernehmen <i>et al.,</i> 2008</a>), so the
| + | src= https://2019.igem.org/wiki/images/e/e9/T--Marburg--ColonyPCRcuredStrain.jpg |
| − | construction of a shuttle-vector based on a modular cloning method significantly improves the genetic toolbox we
| + | alt="gel pcr"> |
| − | created for genetic engineering and synthetic biology approaches in <i>S.elongatus</i> and other
| + | <figcaption> Fig. 12: Colony PCR of the wild type, the conjugated and the cured strain. |
| − | cyanobacteria.<br>
| + | </figcaption> |
| − | </p>
| + | </figure> |
| − | <br>
| + | |
| − | <p style="font-size: 20px">
| + | Our next step was the characterization of the cyanobacterial shuttle vector mentioned in our design section <b>[Link to |
| − | The commonly used <i>S.elongatus</i> strain PCC7942 carries two endogenous plasmids, the 46,4kb pANL (<a
| + | design]</b>. |
| − | href="https://www.ncbi.nlm.nih.gov/pubmed/18353436">Chen <i>et al.,</i> 2008</a>) which is essential and the
| + | In an extensive flow cytometry experiment we assessed the fluorescence of a transformed YFP-construct in our cured |
| − | 7,8kb pANS (<a href="https://www.ncbi.nlm.nih.gov/pubmed/1552863">Van der Plas <i>et al.,</i> 1992</a>) which is
| + | strain, showing that the shuttle vector with the minimal replication element can be maintained in<i>S. elongatus </i> UTEX |
| − | not essential for the strain and can easily be cured.<br>
| + | 2973 . |
| − | This small plasmid has already been used for construction of shuttle vectors (<a
| + | <figure Style="text-align:center"> |
| − | href="https://doi.org/10.1016/0076-6879(87)53054-3">Kuhlemeier Titel anhand dieser DOI in Citavi-Projekt übernehmen & van Arkel, 1987</a> ; <a
| + | <img style="height: 65ex; width: 50ex" |
| − | href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC217787/">Golden & Sherman, 1983</a> ; <a
| + | src= |
| − | href="https://doi.org/10.1099/mic.0.000377">Chen Titel anhand dieser DOI in Citavi-Projekt übernehmen <i>et al.,</i> 2016</a>). <br>
| + | alt="hier kommen noch 3 bilder von FACS Messung aber no time"> |
| − | We followed this lead to create the best shuttle-vector available for cyanobacteria by encompassing the minimal
| + | <figcaption> Fig. 13: hier kommen noch 3 bilder von FACS Messung aber no time. |
| − | replication region of pANS and the ColE1 origin of replication into our vectors, allowing for stable
| + | </figcaption> |
| − | self-replication with high copy numbers in cyanobacteria (<a href="https://doi.org/10.1099/mic.0.000377">Chen Titel anhand dieser DOI in Citavi-Projekt übernehmen
| + | </figure> |
| − | <i>et al.,</i> 2016</a>) and <i>E.coli</i> (<a href="https://doi.org/10.1016/S0065-2660(02)46013-0">Gerhart Titel anhand dieser DOI in Citavi-Projekt übernehmen
| + | After another four weeks of cultivation we looked at our |
| − | <i>et al.,</i>2002</a>). This addition to the genetic toolbox proves invaluable, as it can be easily recovered
| + | cultures again on the UV table to check if fluorescence was still present and the high intensity of the fluorescence |
| − | from the cyanobacterial strain and reintroduced in <i>E.coli</i> for fast GoldenGate-based cloning processes.<br>
| + | proved to us, that the plasmid is still stably replicated in our strain, showing us, that the minimal replication |
| − | </p>
| + | element does indeed work in our strain. |
| − | <br>
| + | For further analysis we performed qPCR with this transformed strain, in order to check the copy number of the |
| − | <p style="font-size: 20px">
| + | vector. We used the copy number of pANL as a reference, which is supposedly at ~2,6 copies per chromosome |
| − | In order to supply the community with an easy selection system, we equipped our shuttle vector with a fluorescent
| + | <a href= https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.000377> (Chen et al., 2016)</a>. Our data shows a ~4,5 times higher copy number relative to pANL, meaning that the construct is |
| − | reporter that is cut out when introducing new genetic parts:<br>
| + | maintained with approximately 11,7 copies per chromosome. |
| − | A mRFP (red fluorescent protein) cassette is flanked by our standardized TypeIIS restriction enzyme recognition
| + | <figure Style="text-align:center"> |
| − | sequences (BsmBI or BsaI depending on what level you want to clone in). In a standard Golden Gate reaction this
| + | <img style="height: 65ex; width: 50ex" |
| − | cassette will drop out and leave space for the parts that should be introduced, allowing for easy selection on
| + | src= |
| − | plate after successful cloning – red colonies are wrong, still harboring the mRFP cassette and white colonies (if
| + | alt="hier kommen noch 3 bilder [FigXX Honrok qPCR data? diagramm biddeeeeeeeeeeeeeee] aber no time"> |
| − | no other fluorescence is introduced) are correct, as the mRFP was switched with the parts of interest.<br>
| + | <figcaption> Fig. 14: hier kommen noch 3 bilder [FigXX Honrok qPCR data? diagramm biddeeeeeeeeeeeeeee] aber no time. |
| − | <br>
| + | </figcaption> |
| − | This crucial part comes in two variations - one for cloning Lvl1 and one for Lvl2 constructs -, giving the Golden
| + | </figure> |
| − | Gate community everything they need for successful and reliable creation of self-replicating vectors in
| + | |
| − | cyanobacteria.
| + | |
| − | </p>
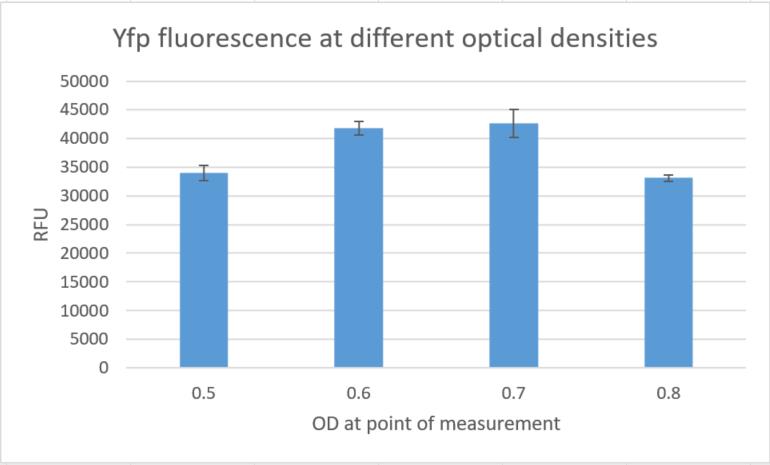
| + | Additionally we measured the fluorescence signals in a plate reader at different optical densities and could again |
| − | <br>
| + | confirm high fluorescence signals, indicating strong gene expression in constructs built around this replication |
| − | </p>
| + | element. |
| − | </div>
| + | <figure Style="text-align:center"> |
| − | <br>
| + | <img style="height: 65ex; width: 50ex" |
| − | </main>
| + | src= https://2019.igem.org/wiki/images/f/f0/T--Marburg--results_yfp_pam_4787_6_replicates.jpg |
| − | | + | alt="diagramm"> |
| | + | <figcaption> Fig. 15:YFP fluorescence at different optical densities. </figcaption> |
| | + | </figure> |
| | + | |
| | + | |
| | + | All this data confirms that the construct actually works and can be reliably used as a cyanobacterial shuttle |
| | + | vector, proving that BBa_K3228069 works as intended, thus functioning as our validated part. |
| | + | This assumption is solidified by all our sequence data, showing that the shuttle vectors were completely assembled |
| | + | as planned in our design section <b>[Link to design of shuttle vectors]</b> |
| | + | . |
| | + | <figure Style="text-align:center"> |
| | + | <img style="height: 65ex; width: 50ex" |
| | + | src= xyz |
| | + | alt="disgramm"> |
| | + | <figcaption> Fig. 16: [FigXX seq results of lvl1 and lvl2 ori] </figcaption> |
| | + | </figure> |
| | + | </p> |
| | + | |
| | + | </p> |
| | </html> | | </html> |
| | <html> | | <html> |