Difference between revisions of "Part:BBa K3078100"
| Line 3: | Line 3: | ||
<h5> | <h5> | ||
<P style="text-indent:2em;"> | <P style="text-indent:2em;"> | ||
| − | This year, a promoter strength measurement device(BBa_K3078100) | + | This year, a promoter strength measurement device(BBa_K3078100) was created by Jilin_China. This part consists of two BbsI digestion sites, RBS and sfGFP. |
</P> | </P> | ||
| Line 9: | Line 9: | ||
<h5> | <h5> | ||
<P style="text-indent:2em;"> | <P style="text-indent:2em;"> | ||
| − | Two | + | Two BbsI recongition sites on the device allow DNA to be constructed on it through GoldenGate Assmbly. And RBS and CDS of sfGFP are at the downstream of the second Bbs I site, which can be used to characterize the promotor intensity by the intensity of fluorescence. // Because the length of promoters in procaryotic organisms are usually short, which means these promoters are easy to be synthesized in vitro. Therefore, we can easily obtain double strand DNA with two sticky ends by single stranded oligonucleotide annealing and we ligand it into the device. This method can construct short promoter measurement parts with in a large quanity and in a short time, which is suitable for the screening and characterizing of promoters. Thus, we consider that this composite part must be popular and widely used in iGEM Community. |
</P> | </P> | ||
<P style="text-indent:2em;"> | <P style="text-indent:2em;"> | ||
| − | This year, we characterized the promoter | + | This year, we characterized the promoter P(sub)59(subed), which hadn't be submitted to iGEM, by the help of this promoter strength measurement device. Compared with 5 promoters from Anderson Family, we identified the relative intensity of P(sub)59(subed) to the control group, attesting this part satisified our anticipation. |
</P> | </P> | ||
<h5> | <h5> | ||
Revision as of 01:46, 22 October 2019
A promoter strength measurement intermediates.
This year, a promoter strength measurement device(BBa_K3078100) was created by Jilin_China. This part consists of two BbsI digestion sites, RBS and sfGFP.
<h5>
Two BbsI recongition sites on the device allow DNA to be constructed on it through GoldenGate Assmbly. And RBS and CDS of sfGFP are at the downstream of the second Bbs I site, which can be used to characterize the promotor intensity by the intensity of fluorescence. // Because the length of promoters in procaryotic organisms are usually short, which means these promoters are easy to be synthesized in vitro. Therefore, we can easily obtain double strand DNA with two sticky ends by single stranded oligonucleotide annealing and we ligand it into the device. This method can construct short promoter measurement parts with in a large quanity and in a short time, which is suitable for the screening and characterizing of promoters. Thus, we consider that this composite part must be popular and widely used in iGEM Community.
This year, we characterized the promoter P(sub)59(subed), which hadn't be submitted to iGEM, by the help of this promoter strength measurement device. Compared with 5 promoters from Anderson Family, we identified the relative intensity of P(sub)59(subed) to the control group, attesting this part satisified our anticipation.
<h5>
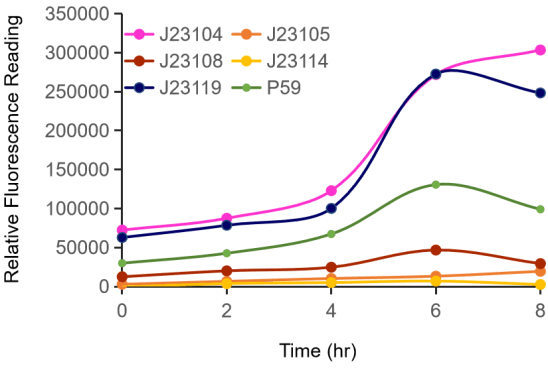
Figure 1. The fluorescence change (Fluorescence/ OD600) of promoters with time. Different promoters were constructed into the same measurement device, cultured over night, diluted to OD600=0.02 and then measured the fluorescence at 528 nm with 485nm excitation wavelength.
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 468
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
<h5>
Figure 1. The fluorescence change (Fluorescence/ OD600) of promoters with time. Different promoters were constructed into the same measurement device, cultured over night, diluted to OD600=0.02 and then measured the fluorescence at 528 nm with 485nm excitation wavelength.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 468
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]