Difference between revisions of "Part:BBa K3254002"
Zongyeqing (Talk | contribs) (→Visual Result as a Normally Closed Switch) |
Zongyeqing (Talk | contribs) (→Results) |
||
| Line 19: | Line 19: | ||
*We can't observed the GFP fluorescence from the experimental tube.<br> | *We can't observed the GFP fluorescence from the experimental tube.<br> | ||
| − | [[File:T--GENAS_China--primary_screening.png|600px|thumb|center| ]]<br> | + | [[File:T--GENAS_China--primary_screening.png|600px|thumb|center|Visual Results as Normally Open Switches]]<br> |
==Quantitative Characterizaion of the Normally Open Switch== | ==Quantitative Characterizaion of the Normally Open Switch== | ||
Revision as of 02:36, 21 October 2019
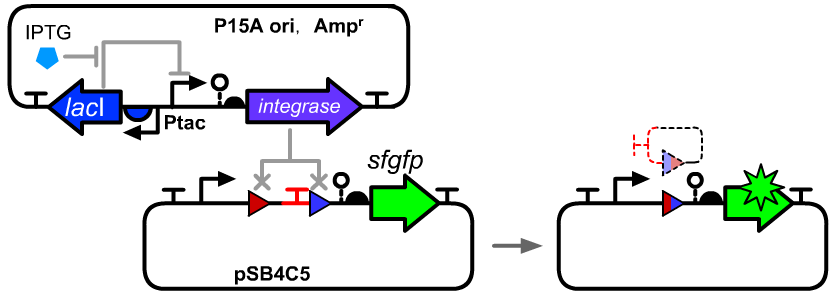
Int7attB-BsaI sites-terminator-Int7attP
We expected that this part can be placed between a promoter and a translational unit part and work as a normally open (NO) switch for the downstreamed gene. and switch to ON state by excising the terminator ECK120033737 between the att sites when it reacted with Int7 integrase (BBa_K3254007). However, we haven't observed the expression of downstreamed gfp gene. That's may caused by the potential terminator activity of attL site after the recombination. Two BsaI restriction site were added between attB site and terminator. As a result, it can work as a normally closed (NC) switch for the gene which was inserted between the two BsaI site and switch to OFF state when it was excised.
Usage and Biology
Visual Result as a Normally Open Switch
- We conducted a simple test to see if our design met the expection.
Experimental Setup
- Genetic design principle of the experimental group is described on the page of BBa_K3254027.
- A P15A-AmpR plasmid was co-transfered into the E.coli DH5α host cell with the reporter plasmid containing this part as the negative control.
- Single colonies were selected from the experimental LB-agar plate and negative control LB-agar plate, then inoculated into EP tubes with 500 μL M9 supplemented medium containing 500 μM IPTG for overnight growth at 37 °C and 200 rpm.
- Tubes were centrifuged at 10000g for 1 min. Then observed the GFP fluorescence of the cell precipitations under blue light.
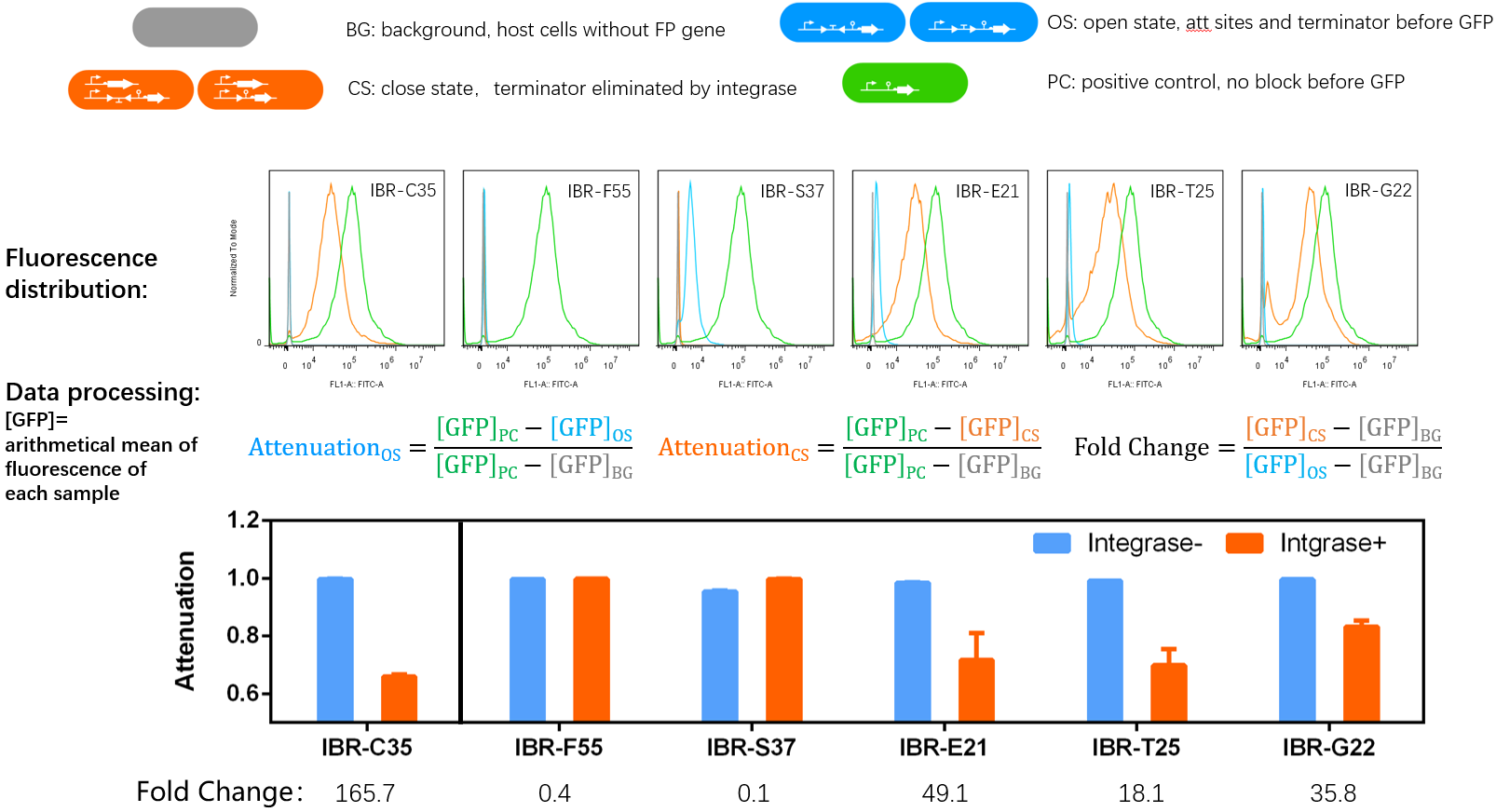
Results
- IBR-C35/F55/S37/E21/T25/G22 indicate the experimental systems for phiC31/Int5/Int7/Int8/Int10/TG1 respctively.
- We can't observed the GFP fluorescence from the experimental tube.
Quantitative Characterizaion of the Normally Open Switch
Experimental Setup
- Bacteria harboring the circuits (see the top part of the result image) first inoculated from single colonies into a flat-bottom 96-well plate for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, 3-μL samples each culture were transferred to a new 96-well plate containing 200 μL per well of PBS supplemented with 2 mg/mL kanamycin.
- The fluorescence distribution of each sample was assayed using a flow cytometry. The arithmetical mean of each sample was determined using FlowJo software.
- The principle of data processing is shown on the result image.
Results
- IBR-C35/F55/S37/E21/T25/G22 indicate the experimental systems for phiC31/Int5/Int7/Int8/Int10/TG1 respctively.
- Compared to other parts, this part can't function as expected. That might due to the potential terminator signal in the attL (or attR) sequence.
- However, it still has the potential to be used as a normally closed switch.
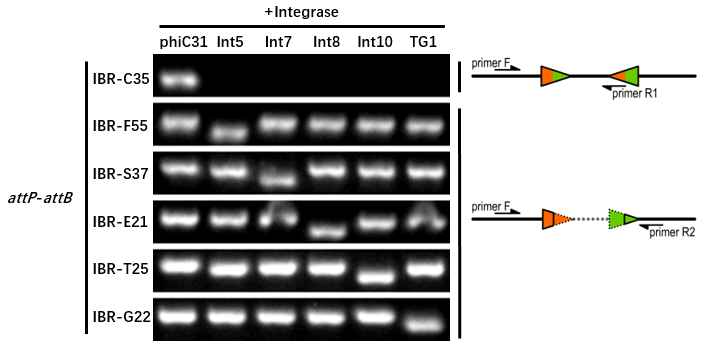
Orthogonality Characterization
Genetic Design
- The composition and principle of the experimental system are indicated below.
Experimental Setup
The reporter plasmid contained this part were co-transferred into E.coli DH5α host with 6 integrase generator plasmids. Then single colonies were inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, The cultures were sampled for genotype PCR testing. The principle of genotype identification was shown on the right of results image.
Results
- IBR-S37 was the plasmid containing this part.
- The result indicates that this part can only be recombined by Int7 integrase.
- The sequence (attL or attR) after recombination should be AGACGAGAAACGTTCCGTCCGTCTGGGTCAGTTGCCTAACCTTAACTTTTACGCAGGTTCAGCTT.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 76
Illegal BsaI.rc site found at 64