Difference between revisions of "Part:BBa K3007021"
| Line 6: | Line 6: | ||
<br> | <br> | ||
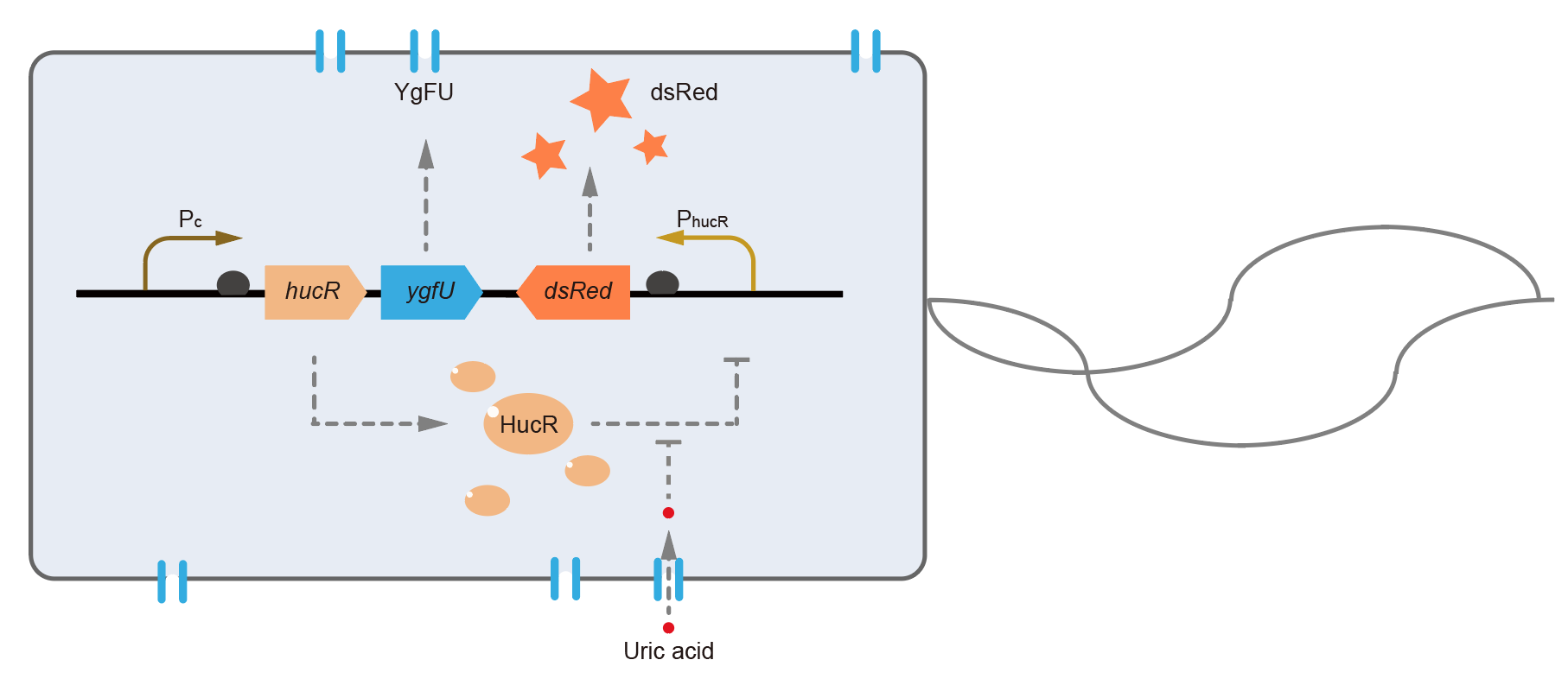
[[File:T--QHFZ-China--BBa K3007001 2.png|400px|thumb|left|Figure 1. Working mechanism of the uric acid detection system in E. coli. ]] | [[File:T--QHFZ-China--BBa K3007001 2.png|400px|thumb|left|Figure 1. Working mechanism of the uric acid detection system in E. coli. ]] | ||
| + | <br> | ||
| + | We used the process shown in Fig. 2 to test if the UA detection system works well. | ||
| + | <br> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 17:54, 20 October 2019
Coding sequence of dsRed
This year, QHFZ-China designed a UA monitor system in E. coli (Fig. 1). Pc is a constitutive promoter, Pcp6 promoter, and it promotes the expression of HucR and YgfU. If the concentration of uric acid (UA) in environment is low, HucR will bind to hucO sequence (located in PhucR), which inhibits the expression of downstream reporter, dsRed or sfGFP. When extracellular UA is present, YgfU can transport UA into the cytoplasm, which leads HucR dissociates from hucO, and induces the fluorescent protein expression.
We used the process shown in Fig. 2 to test if the UA detection system works well.
Sequence and Features
Assembly Compatibility:
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 337
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 337
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 337
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 337
- 1000COMPATIBLE WITH RFC[1000]