Difference between revisions of "Part:BBa K3254018"
Zongyeqing (Talk | contribs) (→Thermodynamic characterization) |
Zongyeqing (Talk | contribs) m |
||
| Line 39: | Line 39: | ||
==Results== | ==Results== | ||
| − | The | + | The results indicate that phiC31 integrase has a good orthogonality with the other 5 att sites and compatible with other integrases. |
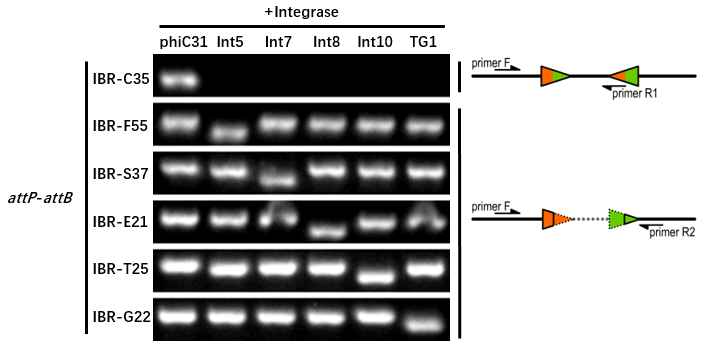
IBR-C35/F55/S37/E21/T25/G22 were the plasmids with phiC31/Int5/Int7/Int8/Int10/TG1 att sites. | IBR-C35/F55/S37/E21/T25/G22 were the plasmids with phiC31/Int5/Int7/Int8/Int10/TG1 att sites. | ||
Revision as of 08:13, 20 October 2019
original sequence of phiC31 integrase gene
This is a conservative design of phiC31 integrase coding sequence. This sequence was isolated from a streptomyces shuttle vector and the EcoRI site was removed. Comparing to BBa_K1039012, this design has a longer coding region, and the "atg" start codon was replaced by "gtg" codon. Because there is an start codon in the upstream, some researchers likely annotated it as a part of the coding sequence. Here we design this part and placed it after a RiboJ(BBa_K1679038) sequence for achieving a appropriate response interval of phiC31 integrase.
Thermodynamic Characterization
- We characterized the thermodynamic characterizatics and orthogonality between phiC31 and 5 other LSTP.
- A new translational unit BBa_K3254010 including this part was constructed.
- The cis-element used in this experiments were BBa_K3254000 which contains the phiC31 att sites.
- We constructed an IPTG inducible system to control the expressional level by the IPTG concentration in E.coli DH5α host.
- The distribution ratios of the cells with original att sites or recombined att sites were measured using a flow cytometry.
Genetic Design
- The composition and principle of the experimental system are indicated below. More details can be seen on the pages of BBa_K3254010 and BBa_K3254000.
- BBa_K3254025 can be seen as a reference of device structure.
Experimental Setup
The two plasmids were co-transformed into E.coli DH5α host. Then we selected colonies with no observable fluoresce and inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with gradient concentrations of IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, 3-μL samples each culture were transferred to a new 96-well plate containing 200 μL per well of PBS supplemented with 2 mg/mL kanamycin. The fluorescence distribution of each sample was assayed using a flow cytometry.
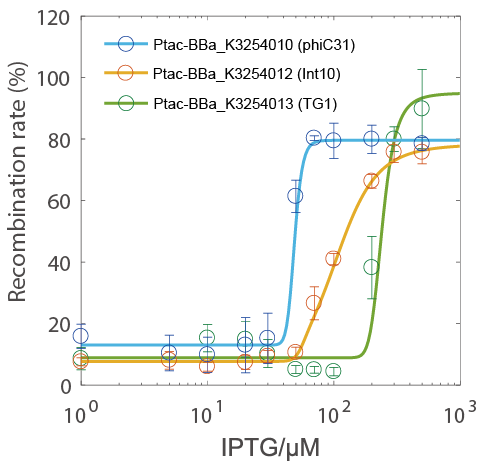
Results
- The recombination rate could be controlled by IPTG concentration and has a very narrow hypersensitivity response interval (the blue circle indicate the phiC31 testing group).
- Because the Ptac promoter has a nearly-linear response curve (see BBa_K3254025), it is more likely that the hypersensitivity is due to the characteristics of the integrase itself rather than the characteristics of the transcriptional regulation system.
Orthogonality Characterization
We conducted orthogonality tests to see the compatibility between the 6 integrase used in our project. The other integrases include Int5, Int7, Int8, Int10 and TG1.
Genetic Design
Similar to the design of thermodynamic characterization. Each integrase generator plasmid were co-transferred with 6 plasmid containing different sets of cis-elements.
Experimental Setup
The growth condition is similar to the thermodynamic characterization but every sample were full induced with 500 μM IPTG. Then we used specific primers to identify the genotype of att sites by PCR. The principle of genotype identification was shown on the right of result image.
Results
The results indicate that phiC31 integrase has a good orthogonality with the other 5 att sites and compatible with other integrases. IBR-C35/F55/S37/E21/T25/G22 were the plasmids with phiC31/Int5/Int7/Int8/Int10/TG1 att sites.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 75
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 1689
Illegal SapI.rc site found at 1380
Illegal SapI.rc site found at 1438
Illegal SapI.rc site found at 1543