Difference between revisions of "Part:BBa K1467400"
(→Calgary 2019) |
|||
| Line 29: | Line 29: | ||
These results indicate that the Golden Gate Module flipper was successful in converting our constructs into a BioBrick compatible composite part, ready to be shipped for the iGEM registry. | These results indicate that the Golden Gate Module flipper was successful in converting our constructs into a BioBrick compatible composite part, ready to be shipped for the iGEM registry. | ||
| − | == [http://2019.igem.org/Team:Calgary Calgary 2019] == | + | == Improvement by [http://2019.igem.org/Team:Calgary Calgary 2019] == |
| + | Group: iGEM Calgary 2019 | ||
| + | Author: Sravya Kakumanu, Sara Far, Nimaya De Silva, Cassandra Sillner | ||
| + | Summary: We improved this part by substituting a strong constitutive promoter [https://parts.igem.org/Part:BBa_J23100 (BBa_J23100)] for the LacI-regulated promoter. We also exchanged the RBS for a stronger one, and codon optimized the part for <i>E. coli</i>. | ||
| + | Documentation: | ||
| + | Part BBa_K2656014 is the monomeric Red Fluorescent Protein 1 coding sequence BBa_E1010 adapted into the Golden Braid assembly method. Thus, this sequence is both compatible with the BioBrick and GoldenBraid 3.0. grammar. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 07:44, 20 October 2019
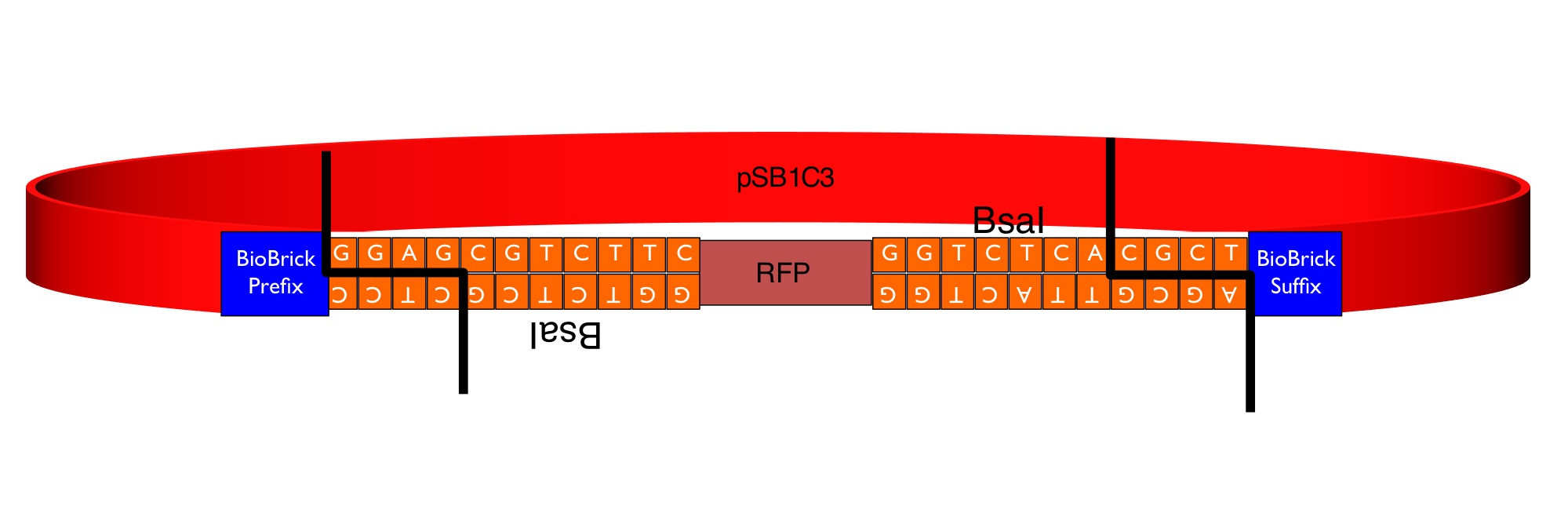
RFP coding device - Golden Gate Module Flipper
This part is a "GoldenGate Flipper" that can be used to assemble complete transcriptional units from GoldenGate MoClo Standard Parts producing a standard BioBrick. It consists of the RFP reporter (from BBa_J04450) flanked by an inverted pair of BsaI recognition sequences. The image below shows the sequences inserted (in orange) between the RFP and the BioBrick prefix and suffix that enable the flipper to accept the parallel assembly of multiple MoClo parts in a one-step digestion-ligation Golden Gate cloning reaction.
The colonies containing the RFP part are clearly red in color under natural light after about 18 hours. Smaller colonies are visibly red under UV. The RFP part does not contain a degradation tag and the RBS is strong. When the flipper reaction is successful the colonies become white as the RFP sequence is replaced by the new part. No part of the BsaI recognition sequence will remain between the BioBrick suffix and the new part.
NRP-UEA 2015
At NRP-UEA we aim to produce a prebiotic of acylated/butrylated starch in plants. To do this, we had to infiltrate our constructs into plants. It was for this reason, along with the fact that we have a range of expertise on site, that we decided to work with GoldenGate cloning. The problem this posed, however, was when we wanted to send our constructs to the iGEM registry at the end of the project, they would be in the wrong backbone and not following the iGEM requirements. For all of our composite parts, we used the GoldenGate Module Flipper to easily convert our constructs into BioBrick compatible composite constructs.
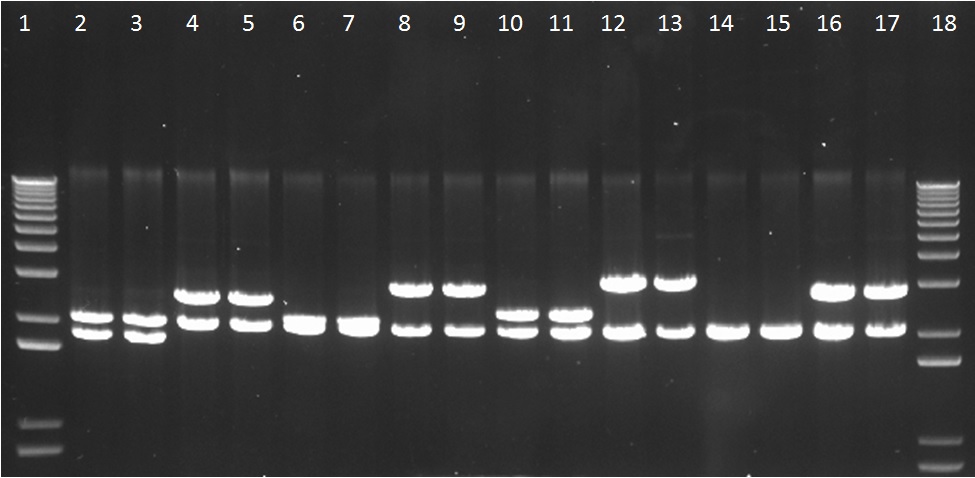
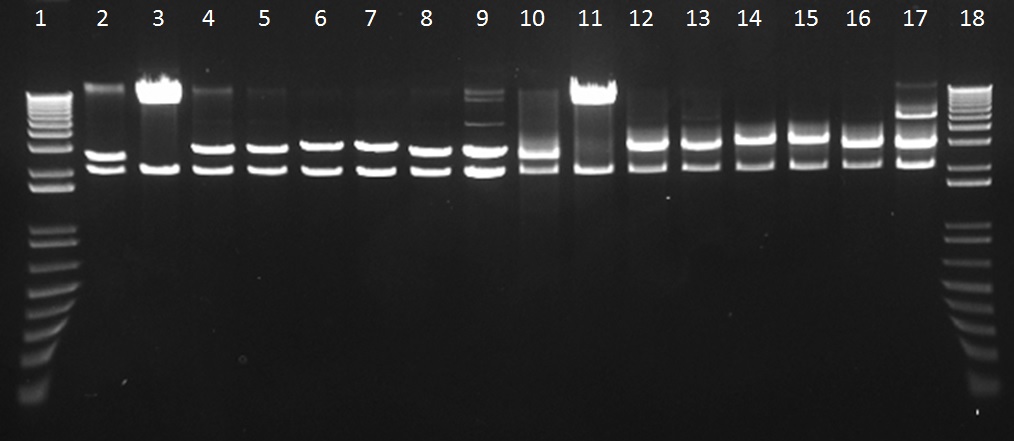
We assembled parts into the MoCloFlipper pSB1C3 according to the GoldenGate one-step Digestion-Ligation. Putative clones were miniprepped and screened by digestion with NotI (Figure 1) and also with Ecor1/PstI(Figure 2) to confirm that cloning was successful and that our constructs had no internal BioBrick restriction sites.
Figure 1: The gel lanes are as follows – 1 = Ladder, 2&3 = BBa_K1618033, 4&5 = BBa_K1618029, 6&7 = BBa_K1618035, 8&9 = BBa_K1618031, 10&11 = BBa_K1618036, 12&13 = BBa_K1618032, 14&15 = BBa_K1618034, 16&17 = BBa_K1618030, 18 = ladder. These indicate that there are no unwanted internal Not1 restriction sites within our constructs.
Figure 2: The gel lanes are as follows – 1 = Ladder, 2&3 = BBa_K1618029, 4&5 = BBa_K1618031, 6&7 = BBa_K1618032, 8&9 = , 10&11 =BBa_K1618029, 12&13 = BBa_K1618031, 14&15 = BBa_K1618032, 16&17 = BBa_K1618030, 18 = Ladder. Lanes 2-9 were digested with Not1, while lanes 10-17 were digested with EcoR1/Pst1. The results indicate that the new tags allow our constructs to be BioBrick compatible.
These results indicate that the Golden Gate Module flipper was successful in converting our constructs into a BioBrick compatible composite part, ready to be shipped for the iGEM registry.
Improvement by [http://2019.igem.org/Team:Calgary Calgary 2019]
Group: iGEM Calgary 2019 Author: Sravya Kakumanu, Sara Far, Nimaya De Silva, Cassandra Sillner Summary: We improved this part by substituting a strong constitutive promoter (BBa_J23100) for the LacI-regulated promoter. We also exchanged the RBS for a stronger one, and codon optimized the part for E. coli.
Documentation: Part BBa_K2656014 is the monomeric Red Fluorescent Protein 1 coding sequence BBa_E1010 adapted into the Golden Braid assembly method. Thus, this sequence is both compatible with the BioBrick and GoldenBraid 3.0. grammar.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 793
Illegal AgeI site found at 905 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1082
Illegal BsaI.rc site found at 6