Difference between revisions of "Part:BBa K2997002"
| Line 3: | Line 3: | ||
<partinfo>BBa_K2997002 short</partinfo> | <partinfo>BBa_K2997002 short</partinfo> | ||
| − | In the human gut microbiome, <i>Clostridium</i> related species have been reported to have the highest conversion capability of fermenting tyrosine into p- | + | In the human gut microbiome, <i>Clostridium</i> related species have been reported to have the highest conversion capability of fermenting tyrosine into <i>p</i>-Cresol<sup>[1]</sup>. To target the root of <i>p</i>-Cresol accumulation, reducing the population of <i>Clostridium</i> is needed. We decide to use <i>C. difficile</i> as a model of <i>p</i>-Cresol producing bacteria because it is a popular research target due to its notorious infectious ability. |
| − | Luckily, iGEM NCKU 2019 was kindly supported by one of our PIs, Professor I-Hsiu, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself in the field of developing novel therapeutic approach for <i>C. difficile</i> infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) from <i>Clostridium Butyricum Miyairi</i> . This bacteriocin was proven to have bactericidal activity against certain strains of <i>Clostridium</i>, including <i>C. difficile</i>[2]. | + | Luckily, iGEM NCKU-Tainan 2019 was kindly supported by one of our PIs, Professor I-Hsiu, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself in the field of developing novel therapeutic approach for <i>C. difficile</i> infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) from <i>Clostridium Butyricum Miyairi</i> . This bacteriocin was proven to have bactericidal activity against certain strains of <i>Clostridium</i>, including <i>C. difficile</i>[2]. |
As proof of concept that bacteriocin is able to inhibit <i>Clostridium</i> growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with <i>C. difficile</i> R20291 strain. | As proof of concept that bacteriocin is able to inhibit <i>Clostridium</i> growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with <i>C. difficile</i> R20291 strain. | ||
| Line 33: | Line 33: | ||
===Reference=== | ===Reference=== | ||
| − | [1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and p- | + | [1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and <i>p</i>-Cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiology Ecology, 94(9). |
[2] Nakanishi, S., & Tanaka, M. (2010). Sequence analysis of a bacteriocinogenic plasmid of Clostridium butyricum and expression of the bacteriocin gene in Escherichia coli. Anaerobe, 16(3), 253–257 | [2] Nakanishi, S., & Tanaka, M. (2010). Sequence analysis of a bacteriocinogenic plasmid of Clostridium butyricum and expression of the bacteriocin gene in Escherichia coli. Anaerobe, 16(3), 253–257 | ||
Revision as of 07:12, 20 October 2019
Bacteriocin short length (sCBM-B)
In the human gut microbiome, Clostridium related species have been reported to have the highest conversion capability of fermenting tyrosine into p-Cresol[1]. To target the root of p-Cresol accumulation, reducing the population of Clostridium is needed. We decide to use C. difficile as a model of p-Cresol producing bacteria because it is a popular research target due to its notorious infectious ability.
Luckily, iGEM NCKU-Tainan 2019 was kindly supported by one of our PIs, Professor I-Hsiu, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself in the field of developing novel therapeutic approach for C. difficile infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) from Clostridium Butyricum Miyairi . This bacteriocin was proven to have bactericidal activity against certain strains of Clostridium, including C. difficile[2].
As proof of concept that bacteriocin is able to inhibit Clostridium growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with C. difficile R20291 strain.
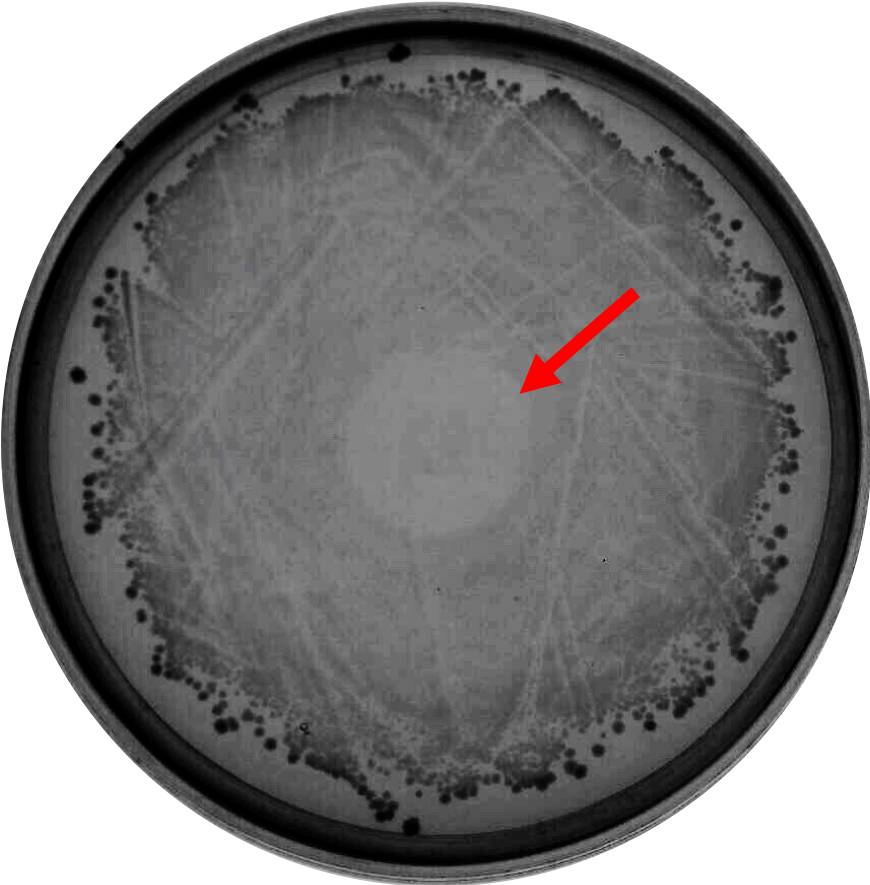
Fig. 1. Spot-on-lawn test using 5 μl purified bacteriocin, inhibition zone formation in the middle of the plate can clearly be seen.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Reference
[1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and p-Cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiology Ecology, 94(9).
[2] Nakanishi, S., & Tanaka, M. (2010). Sequence analysis of a bacteriocinogenic plasmid of Clostridium butyricum and expression of the bacteriocin gene in Escherichia coli. Anaerobe, 16(3), 253–257
