Difference between revisions of "Part:BBa K2997009"
(→Expression in E.coli) |
|||
| Line 17: | Line 17: | ||
Fig. 6. Confirmation of BBa_K2997009 by double digestion, arrow indicates TAL w/ NRBS (~1600bp).M: Marker; Lane 1: pSB1C3-BBa_K2997009; Lane 2: BBa_K2997009 | Fig. 6. Confirmation of BBa_K2997009 by double digestion, arrow indicates TAL w/ NRBS (~1600bp).M: Marker; Lane 1: pSB1C3-BBa_K2997009; Lane 2: BBa_K2997009 | ||
| + | |||
| + | ===RT-PCR=== | ||
| + | RT-PCR experiment was used to confirm that the constructed TAL Biobrick is being transcribed. As seen in Fig.9, cDNA for both bacteria carrying TAL constructs are being detected by PCR. Confirming that the TAL genes is actually being transcribed in E. coli Nissle. | ||
| + | <html> | ||
| + | <br> | ||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/6/62/T--NCKU_Tainan--RT-PCR.png" style="width:30%;"> | ||
| + | </div> | ||
| + | <br> | ||
| + | </html> | ||
| + | <html> | ||
| + | <br> | ||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/2/21/T--NCKU_Tainan--TAL_RT-PCR.jpg" style="width:30%;"> | ||
| + | </div> | ||
| + | <br> | ||
| + | </html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 17:44, 19 October 2019
J23100-NRBS-sam8 (J23100-I742106)
Background
Tyrosine ammonia-lyase (TAL) is an enzyme that converts tyrosine into p-Coumaric acid. We engineered E. coli Nissle to express TAL to turn the excess tyrosine inside the gut into p-Coumaric acid, and compare the different constructs for improvement, BBa_K2997009 and BBa_K2997010.
Expression in E.coli
We constructed Tyrosine Ammonia Lyase with a native RBS , and transformed the plasmid into E. coli Nissle 1917 and confirmed it by double digestion. The results are as follows:
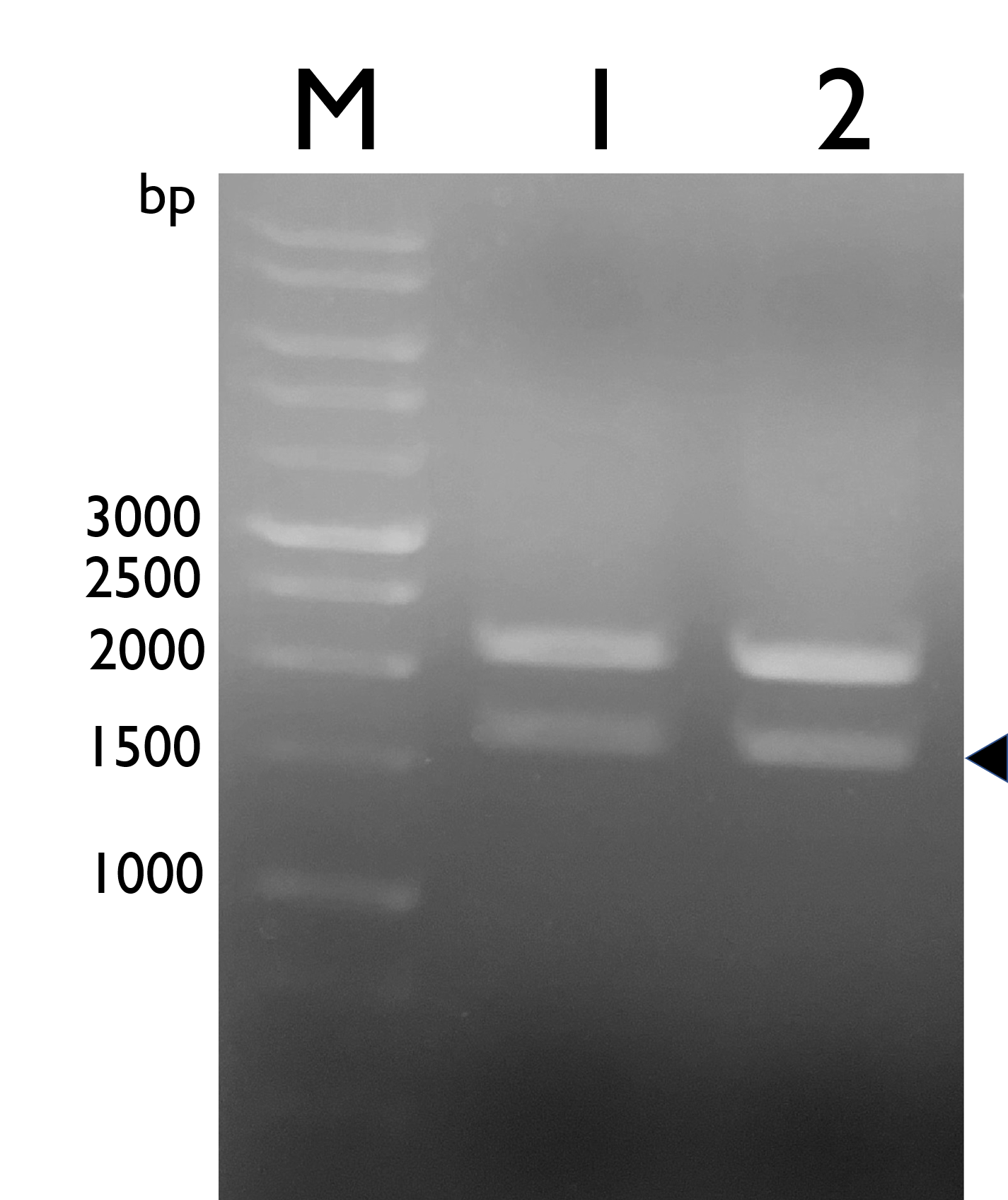
Fig. 6. Confirmation of BBa_K2997009 by double digestion, arrow indicates TAL w/ NRBS (~1600bp).M: Marker; Lane 1: pSB1C3-BBa_K2997009; Lane 2: BBa_K2997009
RT-PCR
RT-PCR experiment was used to confirm that the constructed TAL Biobrick is being transcribed. As seen in Fig.9, cDNA for both bacteria carrying TAL constructs are being detected by PCR. Confirming that the TAL genes is actually being transcribed in E. coli Nissle.


Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 457
Illegal BamHI site found at 36 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 232
