Difference between revisions of "Part:BBa K2460007"
Zongyeqing (Talk | contribs) m (→Characterization by GENAS_China 2019) |
Zongyeqing (Talk | contribs) (→Characterization by GENAS_China 2019) |
||
| Line 74: | Line 74: | ||
===Experimental Setup=== | ===Experimental Setup=== | ||
| − | The growth condition is similar to the thermodynamic characterization but every sample were full induced with 500 μM IPTG. Then we used specific primers to identify the genotype of att sites. The principle of genotype identification was shown on the right of result image. | + | The growth condition is similar to the thermodynamic characterization but every sample were full induced with 500 μM IPTG. Then we used specific primers to identify the genotype of att sites by PCR. The principle of genotype identification was shown on the right of result image. |
===Results=== | ===Results=== | ||
The result indicate that TG1 integrase has a good orthogonality with the other 5 att sites and compatible with other integrases. | The result indicate that TG1 integrase has a good orthogonality with the other 5 att sites and compatible with other integrases. | ||
| + | IBR-C35/F55/S37/E21/T25/G22 were the plasmids with phiC31/Int5/Int7/Int8/Int10/TG1 att sites. | ||
[[File:T--GENAS_China--orthogonality_test.png]] | [[File:T--GENAS_China--orthogonality_test.png]] | ||
Revision as of 11:13, 19 October 2019
TG1 integrase
This CDS codes for the steptomyces phage TG1 integrase, which catalyzes a site specific recombination between TG1 attB (BBa_K2460008) and attP (BBa_K2460009) sequences. This recombinase can be used to flip a DNA sequence flanked by attB and attP sites, as well as integrating a circular DNA with a attB/P site into a linear/circular DNA with a attP/B site.
Biological Characterisation
TG1 integrase is a member of the large serine recombinases(LSRs) family. LSRs are recombinases encoded by temperate phage genomes, which precisely cut and recombine DNA in a highly controllable and predictable way. The recombinases recognize and bind to attB/P sites as dimers, and cut and recombinate on the attB/P sites, with two dimers forming a tetramer. The recombinated attB/P sites form attL and attR sites.
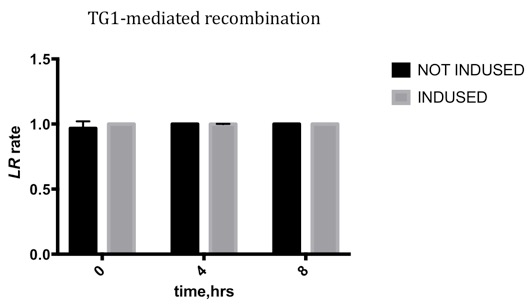
Kinetic Characterization
We conducted qPCR to record the ratio of attL/R to the sum of attL/R and attB/P, to characterize the efficiency varying with time.
Genetic design
- The integrase is expressed under a PBad promoter (BBa_I13453). Considering the leakage of PBad promoter would lead to flipping,the promoter is followed by a weaker RBS (BBa_B0033). Ahead of the promoter,an attB and attP are opposite to each other,flanking a 50 bp random sequence. This construct was cloned on a medium copy plasmid with p15A replication of origin and kanamycin resistance.
- To reduce leakage when not induced, AraC is expressed under a constitutive promoter, cloned on a high copy plasmid with pUC19 replication of origin and Ampicillin resistance.
Experimental Setup
- Since transforming the two plasmid above at a time was unable to prevent the flipping, the plasmid with AraC was first transformed into E.coli DH10b, then the plasmid with the integrase was transformed into the competent cell with the AraC plasmid.
- After culturing for over 2 hours in 2×YT borth, the cells were induced in 1% Arabinose for 8 hours. We sampled the culture at 0 hour, the 4th hour and the 8th hour.
- We calculated flipped and unflipped random sequence by qPCR.
Results
Due to the leakage before inducing, the LR rate changing is not obvious, which means the TG1 integrase is highly efficient, and the leakage may be reduced by improving the genetic design.
Orthogonality Characterization
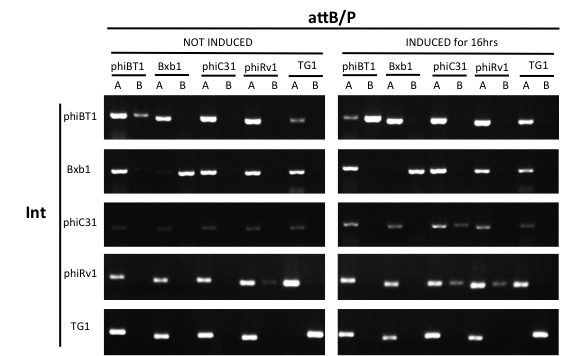
Since our circuit are using multiple integrases, it's important that the attB/P site of one integrase won't be flipped by one another integrase. We conducted orthogonality tests to see the compatibility between our integrases, including phiBT1 (BBa_K2460001), Bxb1 (BBa_K907000), phiC31 (BBa_K1039012), phiRv1 (BBa_K2460004) and TG1 (BBa_K2460007) integrase.
Genetic design
- 5 pairs of attB/P sites are each flanking a 50 bp random sequence, lining up ahead of the PBad promoter (BBa_I13453), an RBS (BBa_B0033) and an integrase. This construct was cloned on a medium copy plasmid with p15A replication of origin and kanamycin resistance.
- The same AraC plasmid in Kinetic Characterization was used in orthogonality Characterization.
Experimental Setup
- The same method of transformation in Kinetic Characterization was used in Kinetic Characterization.
- After culturing for over 2 hours in 2×YT borth, the cells were induced in 1% Arabinose 16 hours.
- Conduct PCR for each integrase's test and control group with primers from 5 pairs of attB/P sites, 2 pairs of primers for each pair of attB/P site.
Results
Expect for the TG1 attB/P site,none of the attB/P sites flipped, indicating that TG1 integrase has a good orthogonality with other attB/P sites and compatible with other integrases.
Characterization by GENAS_China 2019
- Group: GENAS_China 2019
We characterized the thermodynamic characterization and orthogonality between TG1 and 5 other LSTP. A new translational unit BBa_K3254013 including this part was constructed. The cis-element used in this experiments were BBa_K3254005 which contains the TG1 att sitesBBa_K2460008 and BBa_K2460009.
Thermodynamic Characterization
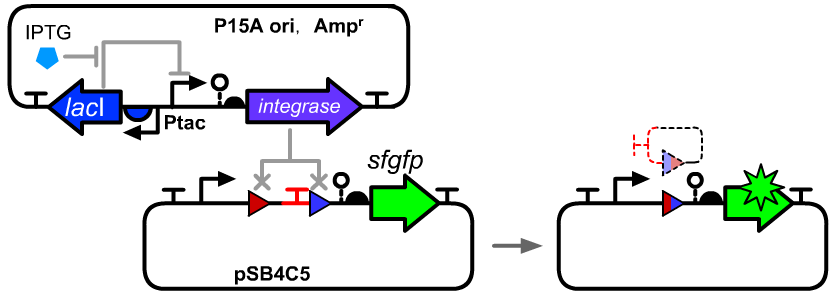
We constructed an IPTG inducible system to control the expressional level by the IPTG concentration in E.coli DH5α host. The distribution ratios of the cells with original att sites or recombined att sites were measured using a flow cytometry.
Genetic design
The composition and principle of the experimental system are indicated below. More details can be seen on the pages of BBa_K3254013 and BBa_K3254005.
Experimental Setup
The two plasmids were co-transformed into E.coli DH5α host. Then we selected colonies with no observable fluoresce and inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, 3-μL samples each culture were transferred to a new 96-well plate containing 200 μL per well of PBS supplemented with 2 mg/mL kanamycin. The fluorescence distribution of each sample was assayed using a flow cytometry.
Results
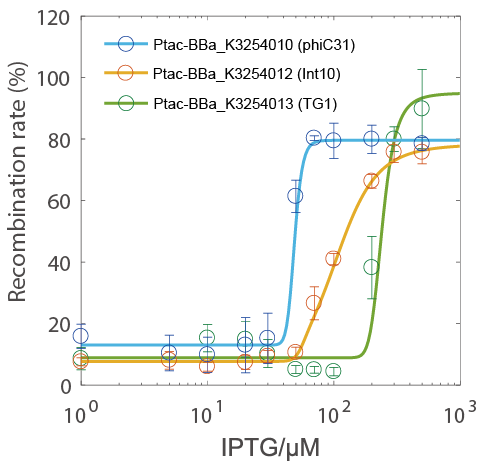
The recombination rate could be controlled by IPTG concentration and has a very narrow hypersensitivity response interval (the green circle indicate the TG1 testing group).
Orthogonality Characterization
We conducted orthogonality tests to see the compatibility between the 6 integrase used in our project. The other integrases include phiC31, Int5, Int7, Int8 and Int10.
Genetic Design
Similar to the design of thermodynamic characterization. Each integrase generator plasmid were co-transferred with 6 plasmid containing different sets of cis-elements.
Experimental Setup
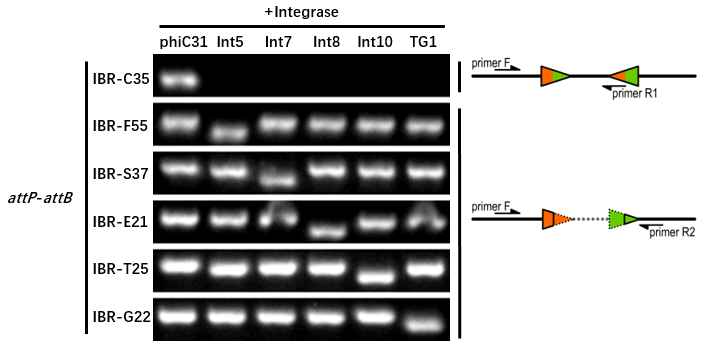
The growth condition is similar to the thermodynamic characterization but every sample were full induced with 500 μM IPTG. Then we used specific primers to identify the genotype of att sites by PCR. The principle of genotype identification was shown on the right of result image.
Results
The result indicate that TG1 integrase has a good orthogonality with the other 5 att sites and compatible with other integrases. IBR-C35/F55/S37/E21/T25/G22 were the plasmids with phiC31/Int5/Int7/Int8/Int10/TG1 att sites.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1725
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1547
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 1761
Illegal SapI.rc site found at 352
Illegal SapI.rc site found at 1513