Difference between revisions of "Part:BBa K1679017"
| Line 22: | Line 22: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| − | <br/> | + | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> |
<br/> | <br/> | ||
<br/> | <br/> | ||
Revision as of 20:12, 15 October 2019
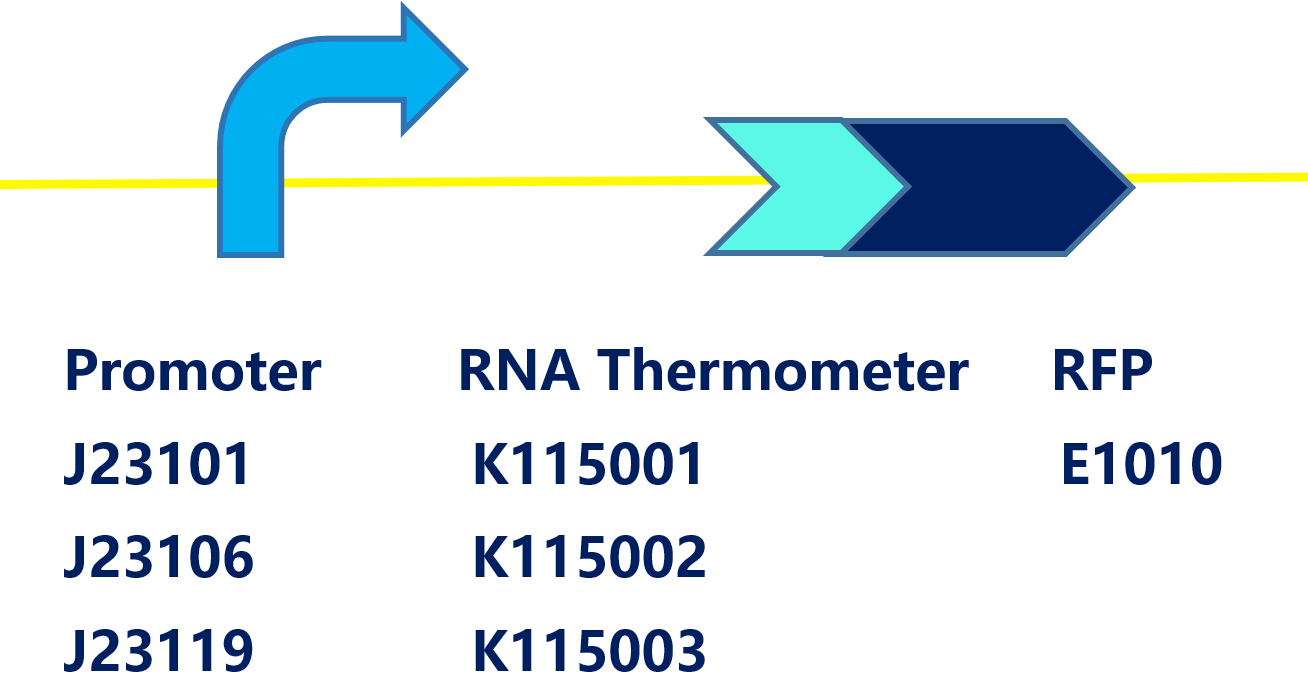
Promoter+RNA thermometer (FourU) +mRFP
This is a device containing a promoter (BBa_J23119),a RNA thermometer (BBa_K115002) and a RFP coding sequence (BBa_E0010) on the pSB1C3, which can be easily transformed into E.coli. RNA Thermometers(RNAT) are temperature-sensing RNA sequences in 5’UTR of their mRNAs. At low temperature, RNAT folds into structure, blocking access of ribosome; At high temperature, RNAT switch from off to open conformation, increasing the efficiency of translation initiation. When the temperature rises above 37°C, RFP would be expressed in theory.
Experiments
TUDelft-2008 has modified 3 kinds of RNAT: ROSE(BBa_K115001), FourU(BBa_ K115002), PrfA(BBa_K115003). We constructed each RNAT under the control of 3 different constitutive promoters:BBa_J23101, BBa_J23106, BBa_J23119 and use RFP as a reporter. After construction, the 9 circuits were transformed into DH5α. According to the results of TUDelft-2008, the temperature threshold is 37℃ for FourU and PfrA, 42℃ for ROSE. Thus, we set the culture temperature to 28℃, 37℃ and 42℃ on solid LB medium while 28℃, 35℃, 37℃, 40℃, and 42℃ in liquid LB medium.
Agar plate test
Liquid test
The 9 circuits were tested through plate reader as well. We set 5 temperature: 28℃, 35℃, 37℃, 40℃ and 42℃. After 41 h, when some replicates had changed color, we calculated the Fluorescence(excitation wavelength-584 nm and emission wavelength-607 nm) and OD(600).
2019 OUC-China's characterization
Background
This part was GFP(mut3b) followed by a variation of the WT SsrA tag sequence, which codes for the amino acid sequence AANDENYNYADAS. When the degradation tag fused to the C-terminal of proteins, it will make the protein susceptible to fast degradation through SspB-mediated binding to the ClpX protease as well as ClpA. DAS variants of SsrA tags require SspB for efficient degradation.
Note that tagged protein degradation rates are dependent on concentration of proteases (ClpX, ClpA) and binding mediators (SspB). Degradation rates are also heavily dependent on protein stability and depend on Km of binding to the protease, which is highly variable with each substrate. Be aware of ClpX, ClpA, and SspB knockouts when using SsrA tags; some expression strains have these mutations.
Result
Compared with GFP(mut3b), we found that the degradation tag has the ability to degrade the protein quickly.
Protocol
Extend to our project
This year, OUC-China has proposed a guideline to construct modular riboswitch for the future iGEM teams. The modular riboswitch we designed consists of the original riboswitch, Stabilizer and Tuner. Stabilizer can protect the structure of riboswitch from damage while Tuner have the ability to separate Stabilizer and GOI. Furthermore, we came up with a idea to help reduce intacellular space utilization. We used a degradation tag to degrade Stabilizer. It has a good result!
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 656
Illegal AgeI site found at 768 - 1000COMPATIBLE WITH RFC[1000]