Difference between revisions of "Part:BBa K2997001"
| Line 3: | Line 3: | ||
<partinfo>BBa_K2997001 short</partinfo> | <partinfo>BBa_K2997001 short</partinfo> | ||
| − | In the human gut microbiome, | + | In the human gut microbiome, <i>Clostridium</i> related species have been reported to have the highest conversion capability of fermenting tyrosine into p-cresol[1]. To target the root of p-cresol accumulation, reducing the population of <i>Clostridium</i> is needed. We decide to use <i>C. difficile</i> as a model of p-cresol producing bacteria because it is a popular research target due to its notorious infectious ability. |
| − | + | ||
| + | Luckily, iGEM NCKU 2019 was kindly supported by one of our PIs, Professor I-Hsiu, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself in the field of developing novel therapeutic approach for <i>C. difficile</i> infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) from <i>Clostridium</i> Butyricum Miyairi. This bacteriocin was proven to have bactericidal activity against certain strains of <i>Clostridium</i>, including <i>C. difficile</i>[2]. | ||
| + | As proof of concept that bacteriocin is able to inhibit <i>Clostridium</i> growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with <i>C. difficile</i> R20291 strain. | ||
| + | |||
| + | <html> | ||
| + | <br> | ||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/2/2e/T--NCKU_Tainan--spot-on-lawn.png" style="width:35%;"> | ||
| + | </div> | ||
| + | <br> | ||
| + | </html> | ||
| + | Fig. 1. Spot-on-lawn test using 5 μl purified bacteriocin, inhibition zone formation in the middle of the plate can clearly be seen. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 18:48, 19 October 2019
Bacteriocin full length (CBM-B)
In the human gut microbiome, Clostridium related species have been reported to have the highest conversion capability of fermenting tyrosine into p-cresol[1]. To target the root of p-cresol accumulation, reducing the population of Clostridium is needed. We decide to use C. difficile as a model of p-cresol producing bacteria because it is a popular research target due to its notorious infectious ability.
Luckily, iGEM NCKU 2019 was kindly supported by one of our PIs, Professor I-Hsiu, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself in the field of developing novel therapeutic approach for C. difficile infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) from Clostridium Butyricum Miyairi. This bacteriocin was proven to have bactericidal activity against certain strains of Clostridium, including C. difficile[2].
As proof of concept that bacteriocin is able to inhibit Clostridium growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with C. difficile R20291 strain.
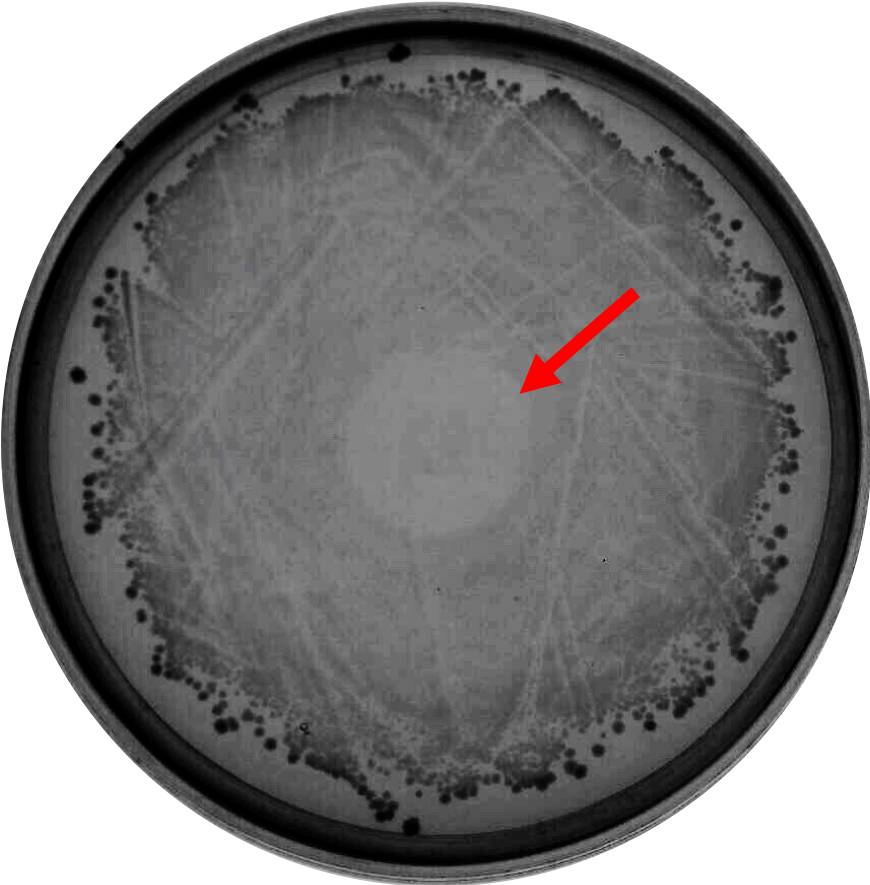
Fig. 1. Spot-on-lawn test using 5 μl purified bacteriocin, inhibition zone formation in the middle of the plate can clearly be seen.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 396
- 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 396
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 396
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 396
- 1000COMPATIBLE WITH RFC[1000]
