Difference between revisions of "Part:BBa K3147004"
(→II. Proof of function) |
|||
| Line 27: | Line 27: | ||
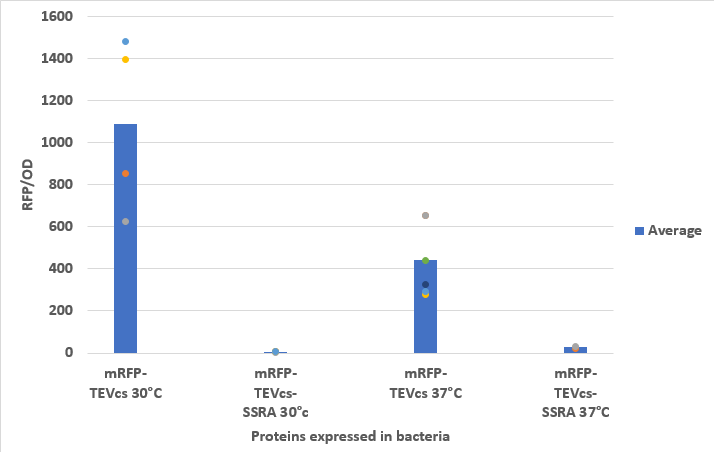
<div align="center"><b>Figure 3</b> :Measurement of the fluorescence at 30°C and 37°C of bacteria expressing mRFP-TEVcs or mRFP-TEVcs-SSRAin RFU</div> | <div align="center"><b>Figure 3</b> :Measurement of the fluorescence at 30°C and 37°C of bacteria expressing mRFP-TEVcs or mRFP-TEVcs-SSRAin RFU</div> | ||
| − | + | ==Reference== | |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 16:11, 14 October 2019
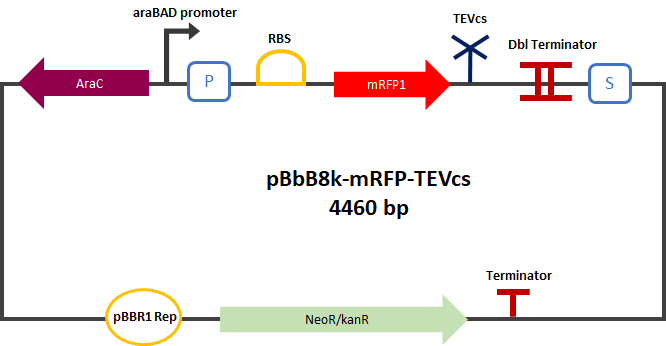
mRFP1 fused to a TEV cleavage site cleaved
I : parts BBa_K3147004 (mRFP1-TEVcs) function
The Montpellier 2019 team made this reporter gene construct in order to compare the basal fluorescence of mRFP1-TEV cutting site with the mRFP1-TEVcs-ssrA (BBa_ K3147003) construct. This construction produces a mRFP1 (BBA_ E1010) fused in C-ter to a TEV cutting site after cutting (ENLYFQ). This construction can be used as a reporter gene.
II. Proof of function
The experimental approach used to test the activity of these reporters was to compare the basal fluorescence rate of this construction to a mRFP1 fused to a ssrA proteolysis tag separated with a TEV recognition site. The construction was cloned by Gibson Assembly in a pBbE8K-RFP (https://www.addgene.org/35363/ ) backbone under the control of a BAD type promoter in order to control its expression.
We compared the basal fluorescence of the strain NEB10β of E. coli transformed with the mRFP1-TEVcs construction to an E. coli NEB10β strain transformed with the mRFP1-TEVcs-ssrA construction. Fluorescence was quantified after arabinose induction at 1% with a plate reader overnight. Here are the fluorescence of mRFP-TEVcs-SSRA and mRFP-TEVcs at 30°C and 37°C.
Reference
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 691
- 1000COMPATIBLE WITH RFC[1000]