Difference between revisions of "Part:BBa I746909"
| Line 3: | Line 3: | ||
This is one of the constructs used to characterise superfolder GFP (see I746916 part description and http://openwetware.org/wiki/IGEM:Cambridge/2008/Improved_GFP for source and other information about this GFP variant) | This is one of the constructs used to characterise superfolder GFP (see I746916 part description and http://openwetware.org/wiki/IGEM:Cambridge/2008/Improved_GFP for source and other information about this GFP variant) | ||
| + | |||
| + | <!-- Below contains the characterization added to this part by TUDelft 2019 --> | ||
| + | |||
| + | <html> | ||
| + | <h2>2019 iGEM team TUDelft</h2> | ||
| + | |||
| + | <p> <a href="https://2019.igem.org/Team:TUDelft">2019 iGEM team TUDelft</a>characterized this part in PURE<sub>frex</sub>.<br><br></p> | ||
| + | |||
| + | |||
| + | </html> | ||
<!-- Below contains the characterization added to this part by Queens_Canada 2019 --> | <!-- Below contains the characterization added to this part by Queens_Canada 2019 --> | ||
Revision as of 12:13, 19 October 2019
superfolder GFP driven by T7 promoter
This is one of the constructs used to characterise superfolder GFP (see I746916 part description and http://openwetware.org/wiki/IGEM:Cambridge/2008/Improved_GFP for source and other information about this GFP variant)
2019 iGEM team TUDelft
2019 iGEM team TUDelftcharacterized this part in PUREfrex.
Characterization: PR4 vs. T7
Group: Queens_Canada, 2019
Author: Ruben Warkentin
Summary: We compared the amount of GFP expressed under a constitutive promoter (medium promoter, strong RBS) to T7 expression. Some proteins fold better under constitutive promoters; however, nobody had yet directly compared the amount of protein produced between constitutive vs. T7 expression.
Methods
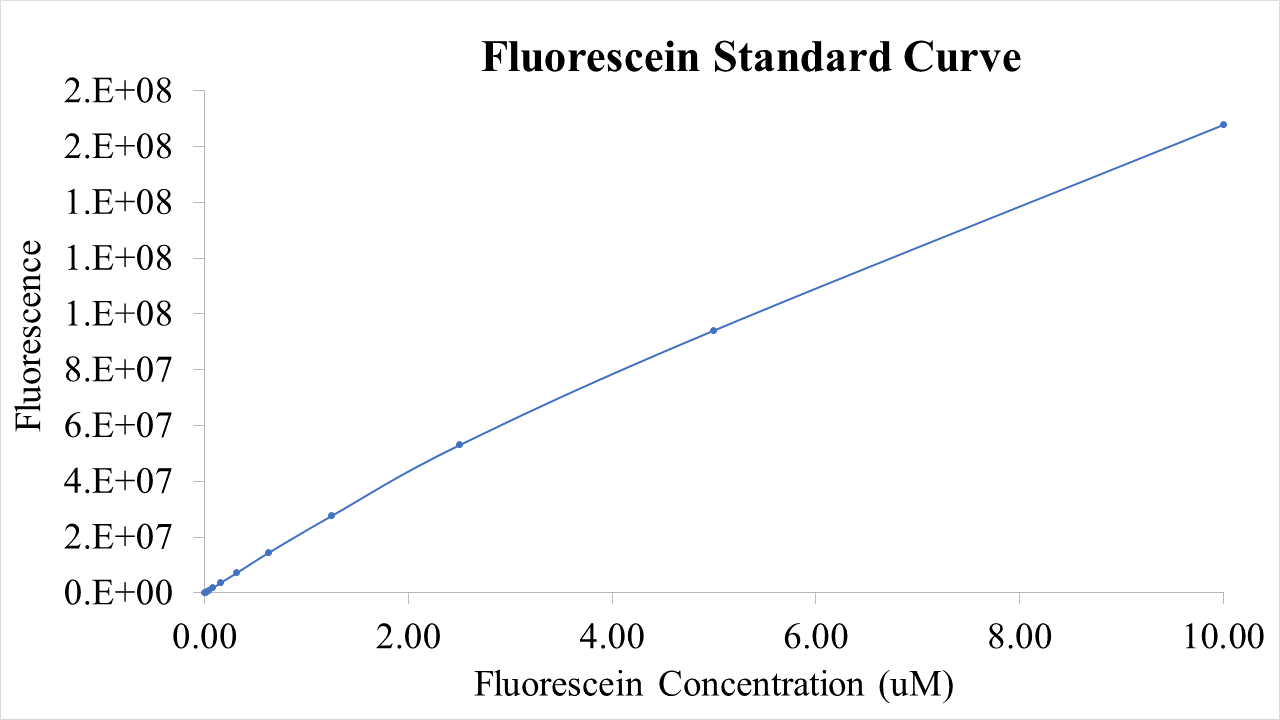
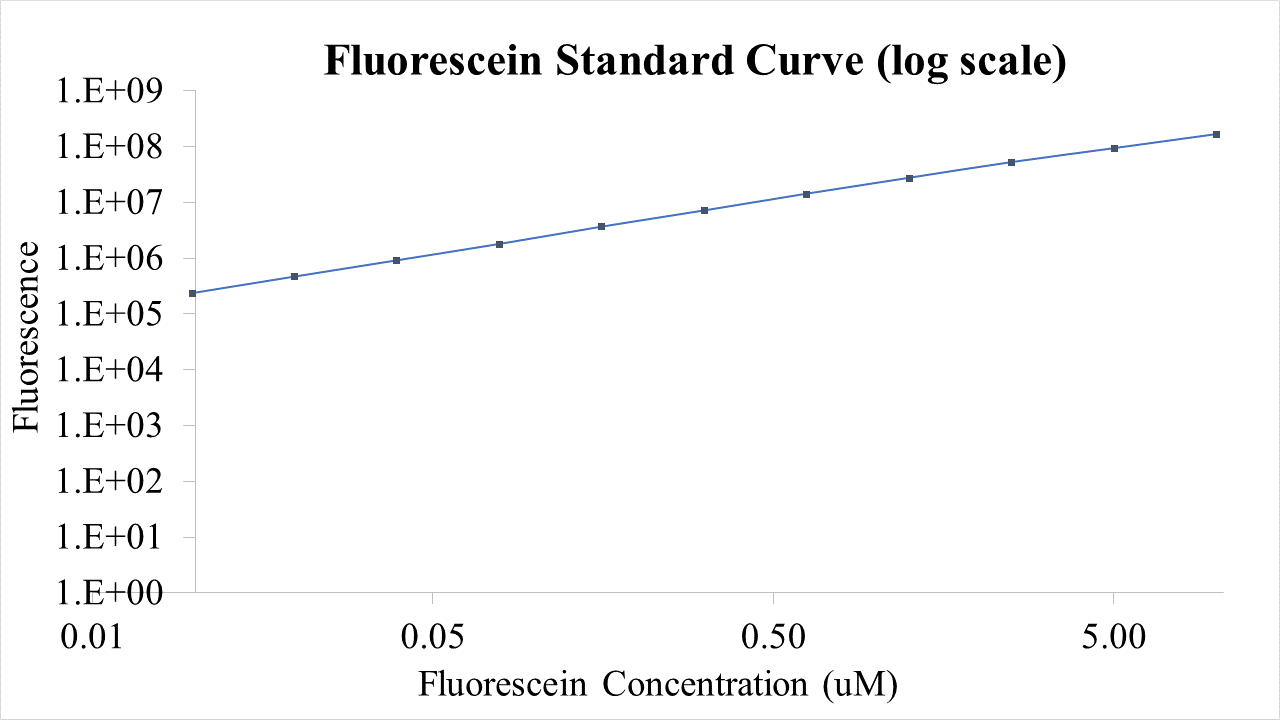
BioBricks were transformed and expressed in E. coli (BL21). BL21 cells were cultured to an OD600=0.6 and 100 uL of culture was transferred into a 96 well plate. Colonies were transfered in quadruplicate. The fluorescence intensity of GFP was measured with a multi-mode microplate reader. The iGEM standardized fluorescence protocol was used for fluorescence measurement standardization (https://www.protocols.io/view/calibration-protocol-plate-reader-fluorescence-cal-6zrhf56).
Results
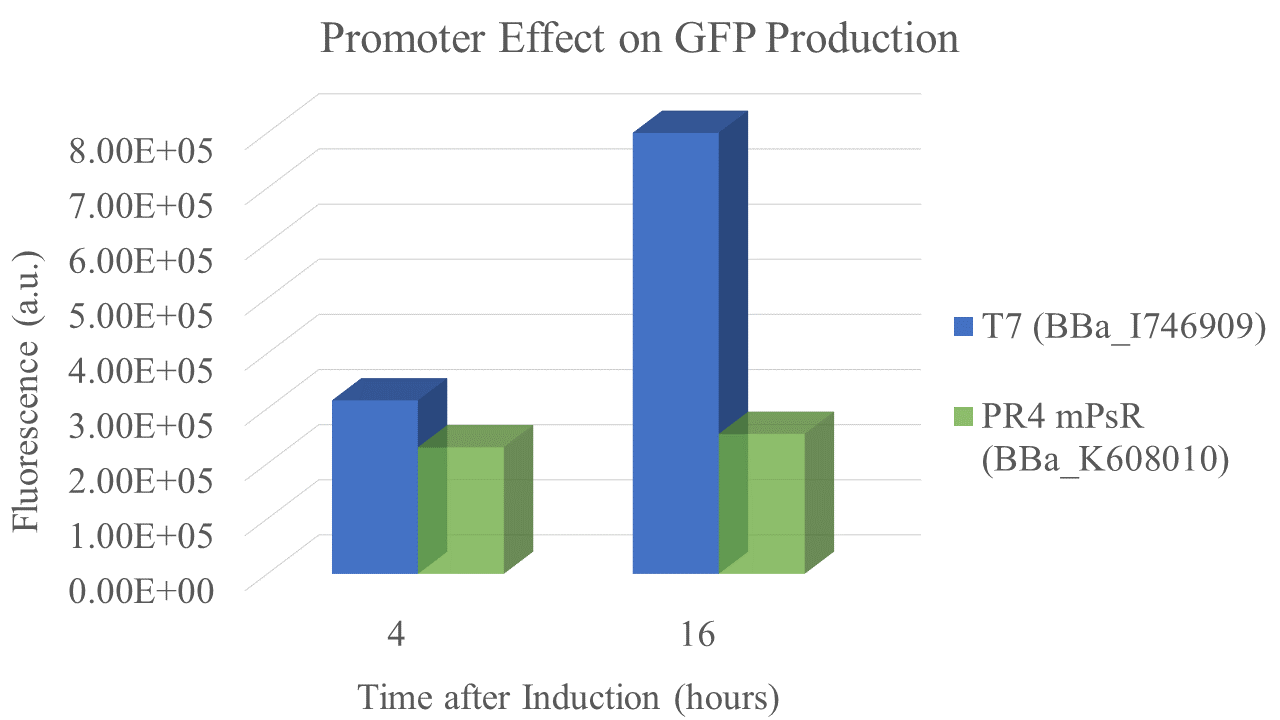
We found that the T7 promoter produced about 2.6 times as much fluorescent signal as the constitutive PR4 promoter, indicating that T7 is much more efficient at producing GFP (Fig. 5). Interestingly, the production of GFP under PR4 did not increase beyond the level observed at 4 hours after inoculation. It seems that PR4 leads to an initial production of protein; however, after the initial expression the promoter seems to be shut off.
Promoter and RBS:
PR4: medium Promoter (J23110) strong RBS (B0034)
T7: T7 Promoter (BBa_I746909)
| sample | PR4 | T7 |
| Fluorescence 4 hrs (a.u.) | 2.24E+05 | 3.09E+05 |
| Fluorescence 16 hrs (a.u.) | 2.48E+05 | 8.00E+05 |
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 62