Difference between revisions of "Part:BBa K3128013"
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K3128013 short</partinfo> | <partinfo>BBa_K3128013 short</partinfo> | ||
| + | |||
| + | OmpX is an outer membrane protein with the C- and N-termini in the intracellulair domain. (<partinfo>BBa_K1761000</partinfo>) | ||
iGEM Grenoble designed this biobrick in order to add a leucine zipper to the C-terminal end of OmpX. | iGEM Grenoble designed this biobrick in order to add a leucine zipper to the C-terminal end of OmpX. | ||
| + | |||
| + | == Usage and Biology == | ||
| + | This description can be found in the <partinfo>BBa_K1761000</partinfo> page. | ||
| + | OmpX, or Outer Membrane Protein X, belongs to a family of highly conserved bacterial proteins that promote bacterial adhesion to and entry into mammalian cells. It presents both C- and N-termini in the intracellular domain (see Figure 1). OmpX consists of an eight-stranded antiparallel all-next-neighbor β barrel. The core of the protein consists of an extended hydrogen-bonding network of highly conserved residues (see Figure 1). [1] | ||
| + | [[File:TU Eindhoven design ompxstructure.png]] | ||
| + | |||
| + | ''Figure 1: A) The OmpX protein structure has been elucidated through NMR and X-ray crystallography, B) The square residues are important for the secondary structure of OmpX. To keep the structure intact, we introduce an amber stop codon in one of the protruding loops. Figure 5B is adapted from [1].'' | ||
| − | + | == Gene Design == | |
| − | == | + | OmpX was identified and characterized by Mecsas et al. and is 513 bp long. It contains a putative signal sequence, which makes sure that OmpX will be in the membrane. [2] |
| + | To be able to create <partinfo>BBa_K3128022</partinfo>, <partinfo>BBa_K3128023</partinfo>, <partinfo>BBa_K3128026</partinfo>, <partinfo>BBa_K3128027</partinfo>, iGEM Grenoble-Alpes team 2019 had to delete the putative signal sequence present in OmpX gene, in order to add it before the leucine zipper gene. In deed, the signal peptide has a conformation enabling the protein addressing to the membrane. This sequence is cut after the translocation. If the OmpX gene is used without changes, the leucine zipper gene is present before the signal peptide, thus generating a cut in the fusion protein leucine-zipper-OmpX. | ||
| − | |||
| − | |||
| − | |||
| + | == References == | ||
| + | [1] J. Vogt and G. E. Schulz, “The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence.,” Structure, vol. 7, no. 10, pp. 1301–9, Oct. 1999. | ||
| − | + | [2] J. Mescas, R. Welch, J.W. Erickson and C.A. Gross, “Identification and characterization of an Outer Membrane Protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including ail of Yersinia enterocolitica.,” Journal of Bacteriology, vol. 177, no. 3, pp. 799-804, Nov. 1994. | |
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 09:29, 23 September 2019
COMP gene without signal peptide
OmpX is an outer membrane protein with the C- and N-termini in the intracellulair domain. (BBa_K1761000)
iGEM Grenoble designed this biobrick in order to add a leucine zipper to the C-terminal end of OmpX.
Usage and Biology
This description can be found in the BBa_K1761000 page.
OmpX, or Outer Membrane Protein X, belongs to a family of highly conserved bacterial proteins that promote bacterial adhesion to and entry into mammalian cells. It presents both C- and N-termini in the intracellular domain (see Figure 1). OmpX consists of an eight-stranded antiparallel all-next-neighbor β barrel. The core of the protein consists of an extended hydrogen-bonding network of highly conserved residues (see Figure 1). [1]
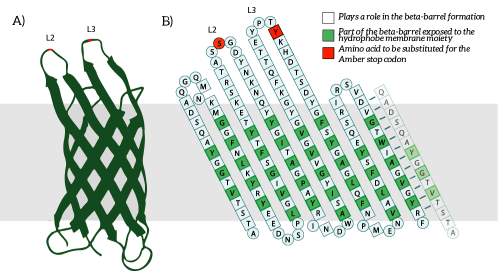
Figure 1: A) The OmpX protein structure has been elucidated through NMR and X-ray crystallography, B) The square residues are important for the secondary structure of OmpX. To keep the structure intact, we introduce an amber stop codon in one of the protruding loops. Figure 5B is adapted from [1].
Gene Design
OmpX was identified and characterized by Mecsas et al. and is 513 bp long. It contains a putative signal sequence, which makes sure that OmpX will be in the membrane. [2] To be able to create BBa_K3128022, BBa_K3128023, BBa_K3128026, BBa_K3128027, iGEM Grenoble-Alpes team 2019 had to delete the putative signal sequence present in OmpX gene, in order to add it before the leucine zipper gene. In deed, the signal peptide has a conformation enabling the protein addressing to the membrane. This sequence is cut after the translocation. If the OmpX gene is used without changes, the leucine zipper gene is present before the signal peptide, thus generating a cut in the fusion protein leucine-zipper-OmpX.
References
[1] J. Vogt and G. E. Schulz, “The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence.,” Structure, vol. 7, no. 10, pp. 1301–9, Oct. 1999.
[2] J. Mescas, R. Welch, J.W. Erickson and C.A. Gross, “Identification and characterization of an Outer Membrane Protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including ail of Yersinia enterocolitica.,” Journal of Bacteriology, vol. 177, no. 3, pp. 799-804, Nov. 1994.
