Difference between revisions of "Part:BBa K3128008"
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | <partinfo> | + | <partinfo>BBa_K3128008 short</partinfo> |
| + | |||
| + | Part from : https://parts.igem.org/Part:BBa_J04450 | ||
monomeric RFP: | monomeric RFP: | ||
Revision as of 14:57, 1 October 2019
Engineered mutant of red fluorescent protein with RFC21 restriction sites
Part from : https://parts.igem.org/Part:BBa_J04450
monomeric RFP: Red Fluorescent Protein. Excitation peak: 584 nm Emission peak: 607 nm
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this protein.
Documentation:
Part BBa_K2656014 is the monomeric Red Fluorescent Protein 1 coding sequence BBa_E1010 adapted into the [http://2018.igem.org/Team:Valencia_UPV/Design Golden Braid assembly method]. Thus, this sequence is both compatible with the BioBrick and GoldenBraid 3.0. grammar.
This coding sequence can be combined with other Golden Braid compatible parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Valencia UPV IGEM 2018 Printeria Collection] to assemble transcriptional units in a one-step BsaI reaction with the [http://2018.igem.org/Team:Valencia_UPV/Protocols Golden Gate assembly protocol].
The characterization of this protein (and by extension of all the other part that codify for the mRFP1) was performed with our transcriptional unit BBa_K2656109. This transcriptional unit was assembled in a GoldenBraid alpha1 plasmid including the following parts:
- BBa_K2656004: the J23106 promoter in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656009: the B0030 ribosome biding site in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- BBa_K2656014: coding sequence
- BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
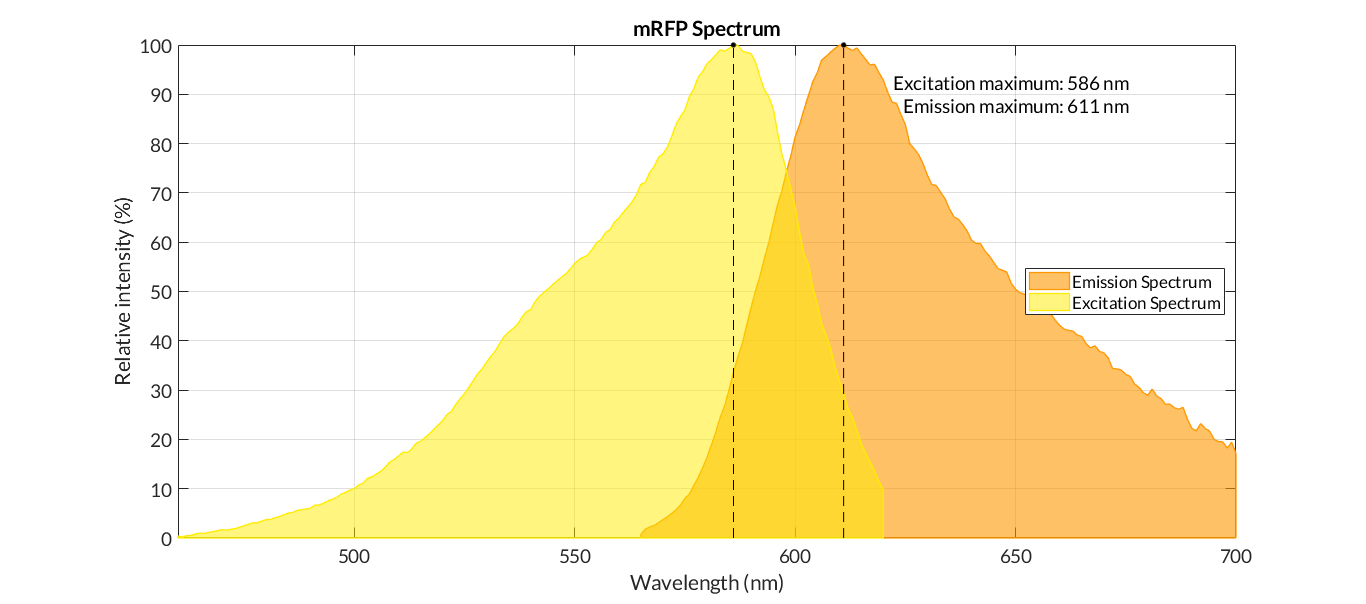
In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we assembled with it, we obtained the emission and excitation spectra in the conditions of our equipment. By using this protocol [http://2018.igem.org/Team:Valencia_UPV/Experiments#spectra] with the parameters of Table 1, Figure 1 was obtained.
| Parameter | Value | ||
| Number of samples | 3 | ||
| Excitation Wavelength measurement range 1 (nm) | [450-620] | ||
| Excitation Wavelength measurement range 2 (nm) | [620-700] | ||
| Emission wavelenght 1 (nm) | 650 | ||
| Emission wavelenght 2 (nm) | 590 | ||
| Emission Wavelength measurement range (nm) | [565-700] | ||
| Excitation wavelenght (nm) | 540 | ||
| Gain (G) | 70 | ||
| Table 1. Parameters used to obtain the spectra | |||
Usage and Biology
Robert E. Campbell started with Discosoma RFP (DsRed) and evolved a faster folding, monomeric variant. See paper listed in source. Codon optimized for expression in bacteria (?? DE)
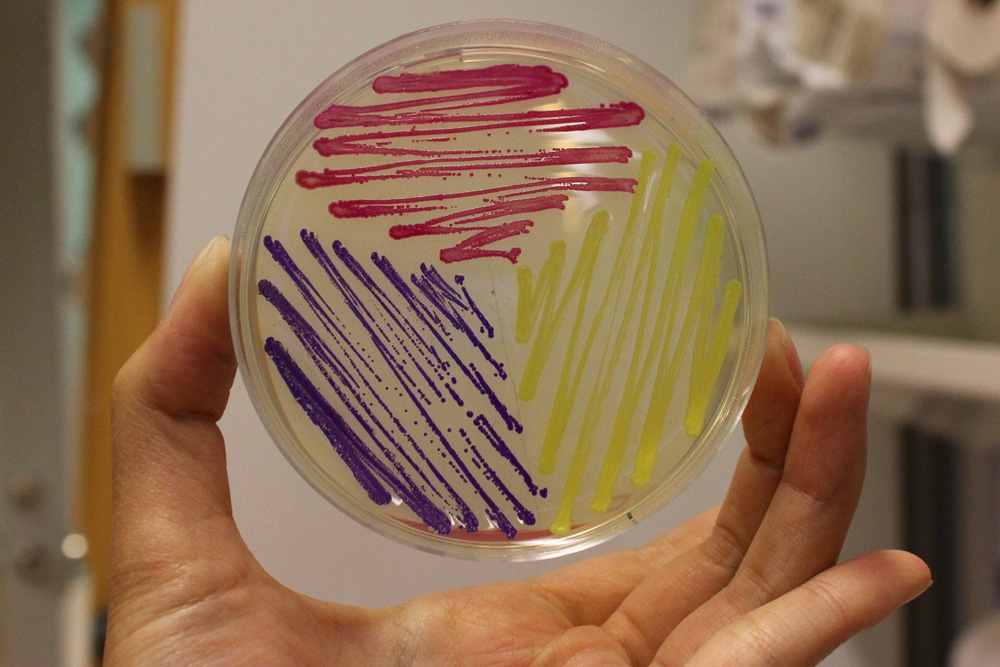
iGEM11_Uppsala-Sweden: Expression of chromoproteins. The images above show E coli No part name specified with partinfo tag. expressing amilCP BBa_K592009 (blue), amilGFP BBa_K592010 (yellow) and RFP BBa_E1010 (red).
Peking iGEM 2016 has fused this part with triple spytag. The fused protein is participate in Peking’s polymer network. By adding this protein, the whole polymer network become visible in most conditions. If you want to learn more about Peking’s polymer network and the role of mRFP in this network, please click here https://parts.igem.org/Part:BBa_K1989004".
Allergen characterization of BBa_E1010: NOT a potential allergen
The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments
For the biobrick part, BBa_E1010, there was a 27.6% of identity match and 56.9% of similarity match compared to the allergen database. This means that the biobrick part is not of potential allergen status. In the 80 amino acid alignments by FASTA, no matches found that are greater than 35% for this biobrick.
>Internal Priming Screening Characterization of BBa_E1010: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Contribution
Group: Hong Kong-CUHK iGEM 2017
Author: Yuet Ching Lin
Summary: We measured the fluorescent signal of mRFP in buffers with different pH.
Documentation:
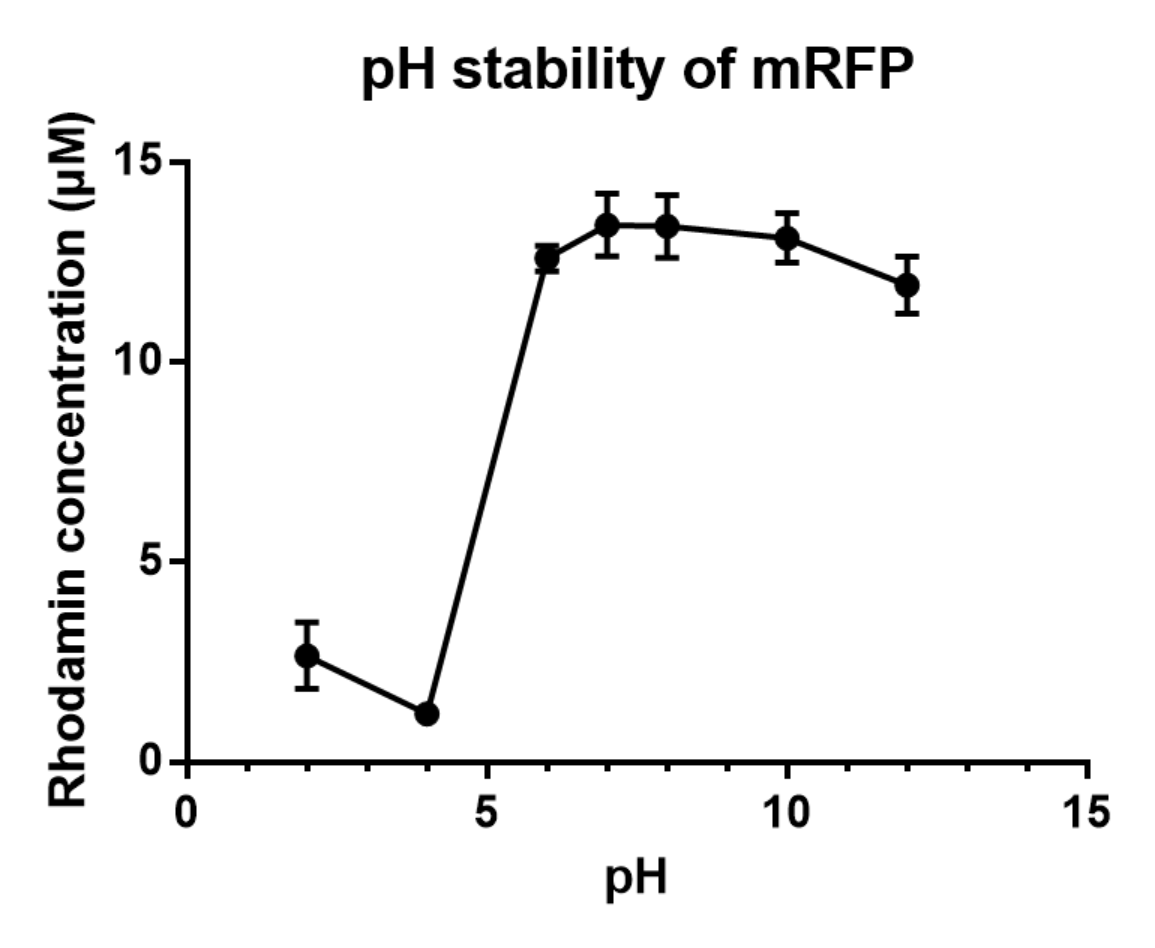
Charaterization of mRFP pH stabillity:
We grew C41 bacteria with parts BBa_J61002 in 2XYT for 24 hours. After purifying the mRFP by Ion Exchange Chromatography and Hydrophobic Interaction Chromatography, we measured the fluoresece (ex ,em ) of purified mRFP, which is diluted to 10µg/100µl (total 200µl) in triplicates, into different buffers (ranges from pH2 to pH12; Volume of mRFP:buffer = 1:1.8). To facilitate reproducibility of the experiment, we correlated the relative fluorescent intensity to an absolute fluorophore concentration by referring it to a standard curve of the fluorophores(Rhodamine) using the interlab study protocol. The result shows that the stability drops dramatically in pH condition below 6 and relatively stable in pH 6-10.
| Measurement Type | Fluorescence |
| Microplate name | COSTAR 96 |
| Scan mode | orbital averaging |
| Scan diameter [nm] | 3 |
| Excitation | 550-20 |
| Emission | 605-40 |
| Dichronic filter | auto 572.5 |
| Gain | 500 |
| Focal height [nm] | 9 |
Contribution2
Part name:BBa_K2382013
Group: iGEM17_CSMU_NCHU_Taiwan 2017
Author: SHAO-CHI LO
Summary: We add a His Tag at the end. Therefore, a fusion protein with this part may have red color and the ability to be purified easily.
Documentation:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 555
Illegal AgeI site found at 667 - 1000COMPATIBLE WITH RFC[1000]
|};
Parts table
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||