Difference between revisions of "Part:BBa K1694003"
| Line 41: | Line 41: | ||
<p style="font-size:120%">'''Improvement by Fudan-CHINA 2018:'''</p> | <p style="font-size:120%">'''Improvement by Fudan-CHINA 2018:'''</p> | ||
| − | In the year 2018, Fudan-CHINA makes new improvement to the VEGF-scFv. In our STEP system, to improve the affinity of the VEGF-scFv to VEGF, we used Rosetta to help design our protein domain. However, we found that the initial version of VEGF-scFv in parts has no accepted structure in PDD (Protein Data Base) of NCBI. So, to improve the reliability of our results, we introduced a kind of VEGF-scFv ([[ | + | In the year 2018, Fudan-CHINA makes new improvement to the VEGF-scFv. In our STEP system, to improve the affinity of the VEGF-scFv to VEGF, we used Rosetta to help design our protein domain. However, we found that the initial version of VEGF-scFv in parts has no accepted structure in PDD (Protein Data Base) of NCBI. So, to improve the reliability of our results, we introduced a kind of VEGF-scFv ([[Part:BBa_K2886002]]) that has accepted structure in PDD to iGEM, making it more accessible to iGEMers. |
<h1>'''Display scFv on the cell surface of ''E. coli'''''</h1> | <h1>'''Display scFv on the cell surface of ''E. coli'''''</h1> | ||
Latest revision as of 19:24, 17 October 2018
Single-chain variable fragment (Anti-VEGF)
Introduction:
ScFv (Single-Chain Variable Fragment)
ScFv (single-chain variable fragment) is a fusion protein containing light (VL) and heavy (VH) variable domains connected by a short peptide linker (Fig. 1). The peptide linker (GGSSRSSSSGGGGSGGGG) is rich in glycine and serine which makes it flexible.
Features of scFv:
1. Specificity:Though remove of the constant regions , scFv still maintain the binding affinity and specificity of the original immunoglobulin.
2. Efficient:ScFv is smaller than the entire antibody, so it place little stress for E. coli to express it
Vascular endothelial growth factor
1. VEGF (Vascular endothelial growth factor), a protein that can stimulates vasculogenesis and angiogenesis. Some cancers can overexpress VEGF, which will cause some vascular disease. Drug bevacizumab can inhibit VEGF and control or slow those diseases.
2. VEGF is a sub-family of growth factors, including VEGF-A, placenta growth factor (PGF), VEGF-B, VEGF-C and VEGF-D.
3. There are three types of VEGF receptors on the cell surface, and VEGFR-2 is the very receptor responses to most VEGF.
Bevacizumab
We selected the single chain variable fragments (scFv) of monoclonal antibodies Bevacizumab and named it anti-VEGF. Bevacizumab is a monoclonal antibody which blocks angiogenesis by inhibiting vascular endothelial growth factor A (VEGF-A). VEGF-A stimulates angiogenesis in a variety of diseases, especially in cancer.
1. When VEGF-A binds to VEGFR-2, it causes two VEGFR-2 combine to form a dimer. This allows for signaling molecules to enter to the cell, bind to the receptor, and become activated. Then start the angiogenesis.
2. Bevacizumab can bind with VEGF released from tumor cell, block VEGFR to inhibit tumor angiogenesis, thereby cutting off the tumor's supplies and prevent tumor from growing.
Reference:
[1]Huston, J. S., Levinson, D., Mudgett-Hunter, M., Tai, M. S., Novotný, J., Margolies, M. N., … Crea, R. (1988). Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 85(16), 5879–5883.
[2]Los, M.; Roodhart, J. M. L.; Voest, E. E. (2007). "Target Practice: Lessons from Phase III Trials with Bevacizumab and Vatalanib in the Treatment of Advanced Colorectal Cancer". The Oncologist 12 (4): 443–50.
[3]Dougher-Vermazen M, Hulmes JD, Böhlen P, Terman BI (November 1994). "Biological activity and phosphorylation sites of the bacterially expressed cytosolic domain of the KDR VEGF-receptor". Biochem. Biophys. Res. Commun. 205 (1): 728–38.</p>
Improvement by Fudan-CHINA 2018:
In the year 2018, Fudan-CHINA makes new improvement to the VEGF-scFv. In our STEP system, to improve the affinity of the VEGF-scFv to VEGF, we used Rosetta to help design our protein domain. However, we found that the initial version of VEGF-scFv in parts has no accepted structure in PDD (Protein Data Base) of NCBI. So, to improve the reliability of our results, we introduced a kind of VEGF-scFv (Part:BBa_K2886002) that has accepted structure in PDD to iGEM, making it more accessible to iGEMers.
Display scFv on the cell surface of E. coli
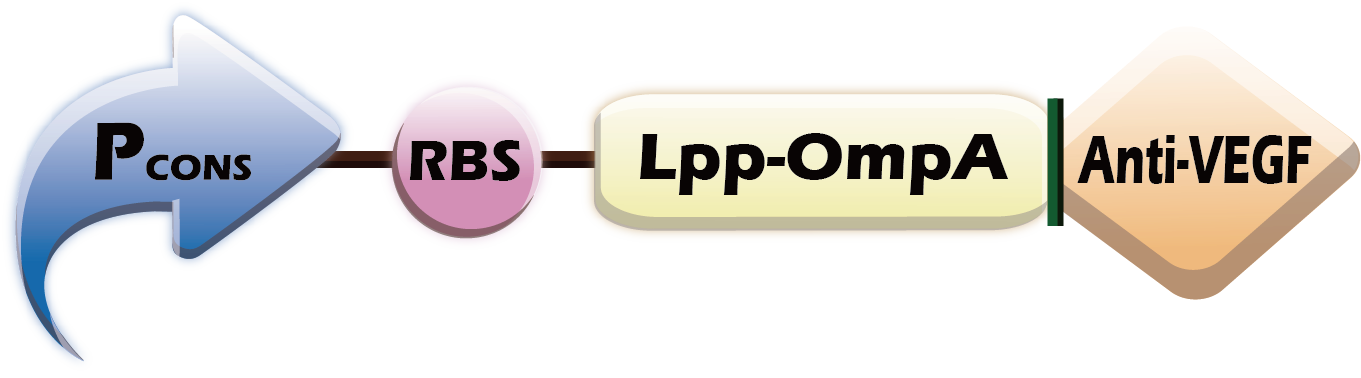
To display the antibody outside the E. coli, we used Lipoprotein-Outer membrane protein A (Lpp-OmpA)
BBa_K1694002. According to the paper reference [1], We chose the first 9 amino acids of Lpp, and the 46~159 amino acids of OmpA.
In order to change the scFv parts easily, we added a NcoI restriction site between OmpA and scFv so that we can change various scFv DNA sequence using the NcoI restriction enzyme.
The (Fig.3) showed how we combine Lpp-OmpA-N and scFv together, first we use restriction enzyme NcoI to digest the upstream and downstream parts. After ligate two digest product, there are no M site in Lpp-OmpA-N-scFv.
See this composite part:BBa_K1694013
Reference:
[1]Improving tumor targeting and therapeutic potential of Salmonella VNP20009 by displaying cell surface CEA-specific antibodies, Michal Bereta, Andrew Hayhurst, Mariusz Gajda, Paulina Chorobik, Marta Targosz, Janusz Marcinkiewicz, Howard L. Kaufman (2007)
Experiment:
After receiving the DNA sequences from the gene synthesis company, we recombined each scFv gene with pSB1C3 backbone and conducted a PCR experiment to check the size of each of the scFvs. The DNA sequence length of the scFvs are around 600~800 bp. In this PCR experiment, the scFv products size should be near at 850~1050 bp. The (Fig.5) showed the correct size of the scFv, and proved that we successful ligated the scFv sequence onto an ideal backbone.
Application of the part:
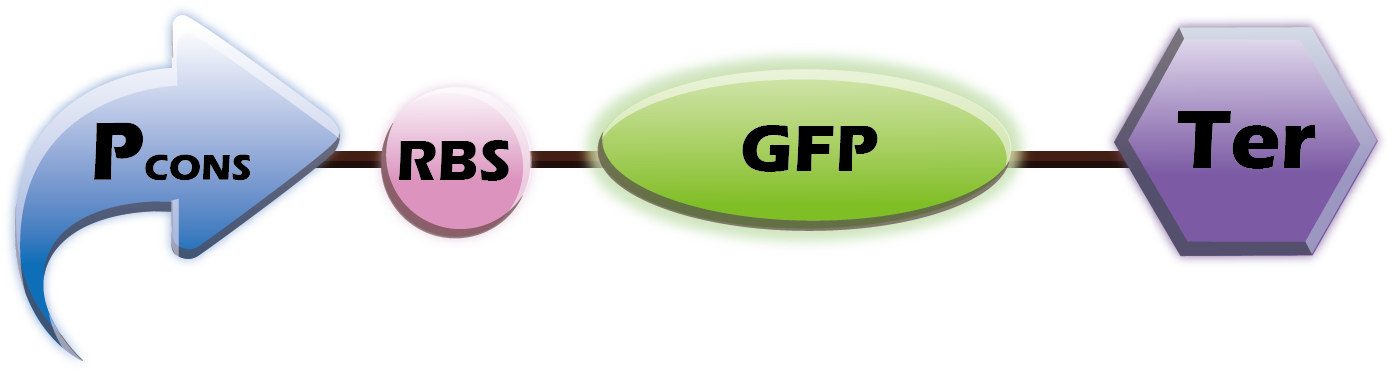
1. Co-transform (Two plasmids)
This year we want to provide a customized platform. We provide two libraries of Pcon+RBS+OmpA-scFv and Pcons+RBS+Fluorescence+Ter into E. coli. Therefore, our customers can choose any scfv and any fluorescence protein. Our team will then co-transform the two plasmids, which helps us tailor our product to the wishes of our customers.
(1) Parts:
(2) Cell staining experiment:
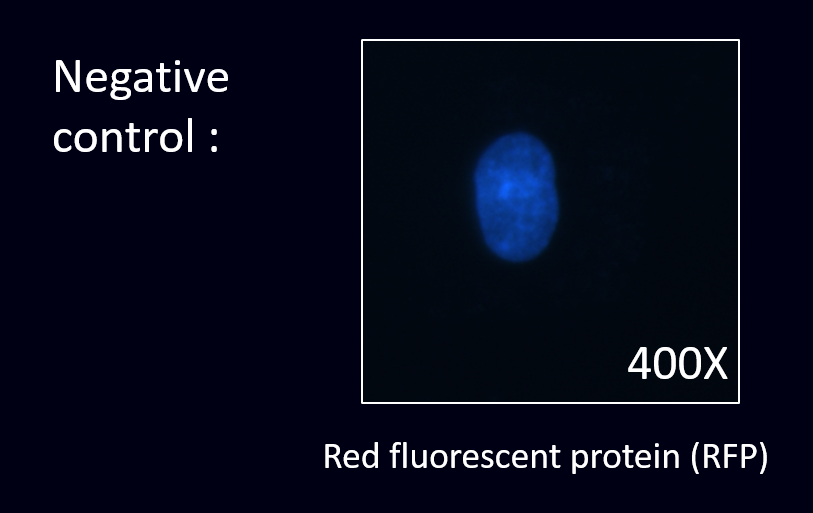
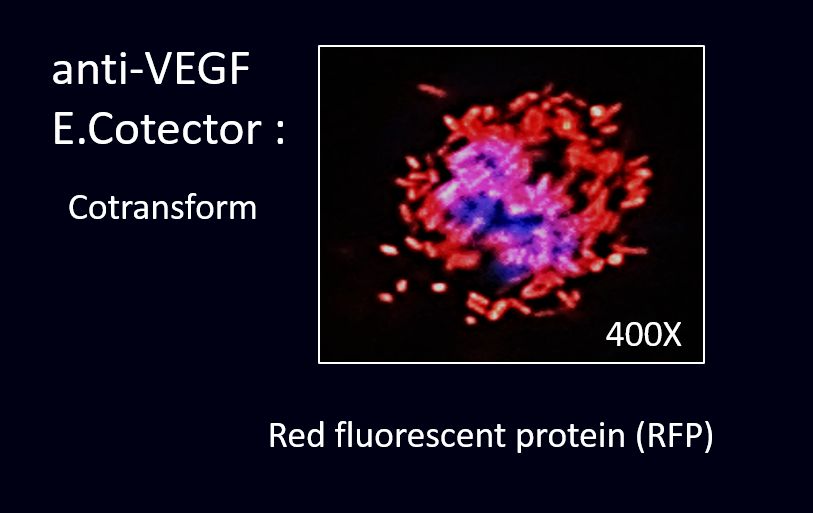
After cloning the part of anti-VEGF, we were able to co-transform anti-VEGF with different fluorescence protein into our E. coli.
The next step was to prove that our co-transformed product have successfully displayed scFv of anti-VEGF and expressed fluorescence protein.
To prove this, we conducted the cell staining experiment by using the co-transformed E. coli to detect VEGF in the cancer cell line.
(3) Staining results:
2. Transformation of single plasmid
To prove that our scFv can actually bind on to the antigen on cancer cells, we connected each scFv with a different fluorescent protein and the blue chromoprotein amilCP. Therefore, we could use fluorescence microscope to clearly observe if the E. coli has produced scFv proteins. Currently, we built three different scFv connected with their respectively fluorescence protein. When applied on cell staining, we can identify the antigen distribution on cancer cells by observing the fluorescence. Furthermore, if we use the three scFv simultaneously, we can also detect multiple markers.
(1) Parts:
(2) Cell staining experiment:
After creating the part of scFv and transforming them into our E. coli, we were going to prove that our detectors have successfully displayed scFv of anti-VEGF. To prove this, we have decided to undergo the cell staining experiment by using our E. coli to detect the VEGF in the SKOV-3 cancer cell lines. SKOV-3 is a kind of epithelial cell that expressed markers such as VEGF.
(3) Staining results:
Modeling
In the modeling part, we discover optimum protein expression time by using the genetic algorithm in Matlab.
We want to characterize the actual kinetics of this Hill-function based model that accurately reflects protein expression time.
When we have the simulated protein expression rate, the graph of protein production versus time can be drawn. Thus, we get the optimum protein expression time.
Compared with the simulated protein production rate of time, our experiment data quite fit the simulation.
Co-transform
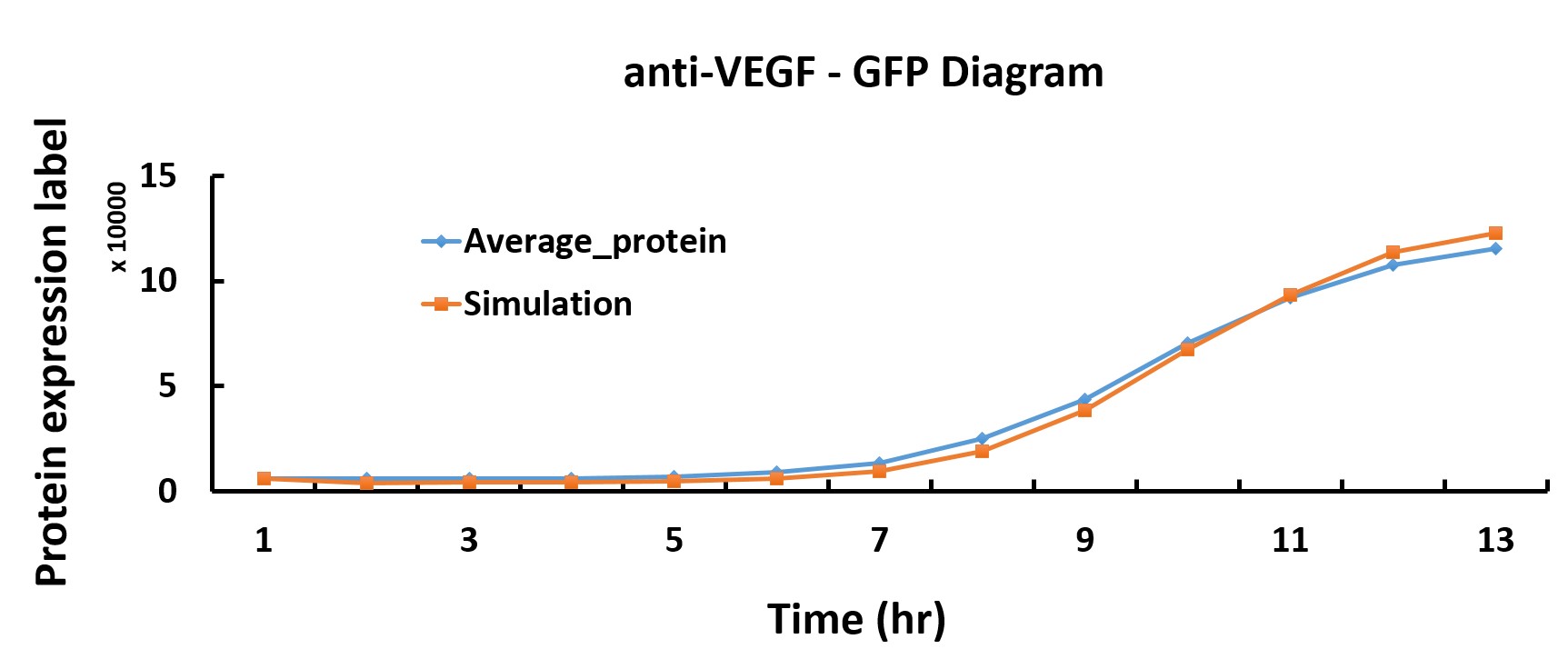

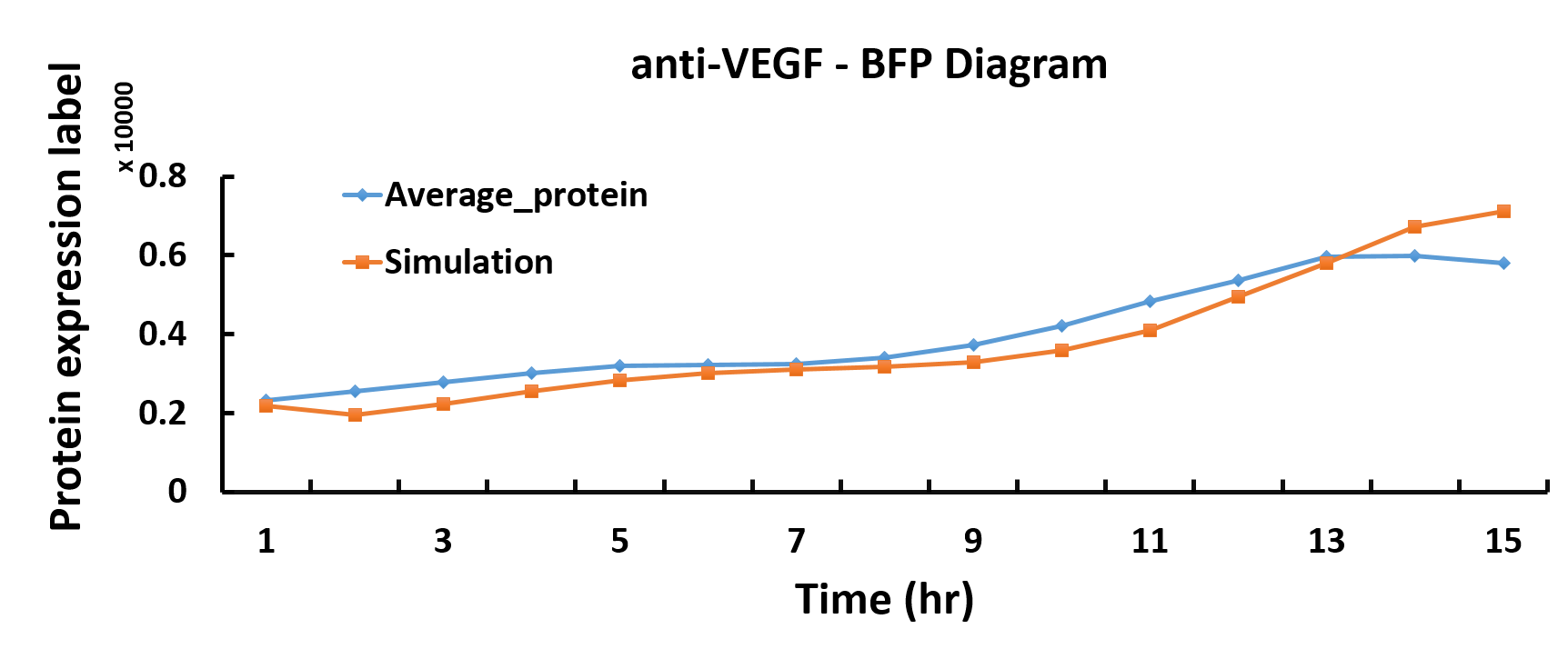
Transformation of single plasmid


Usage and Biology
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]