Difference between revisions of "Part:BBa K2611010"
| Line 3: | Line 3: | ||
<partinfo>BBa_K2611010 short</partinfo> | <partinfo>BBa_K2611010 short</partinfo> | ||
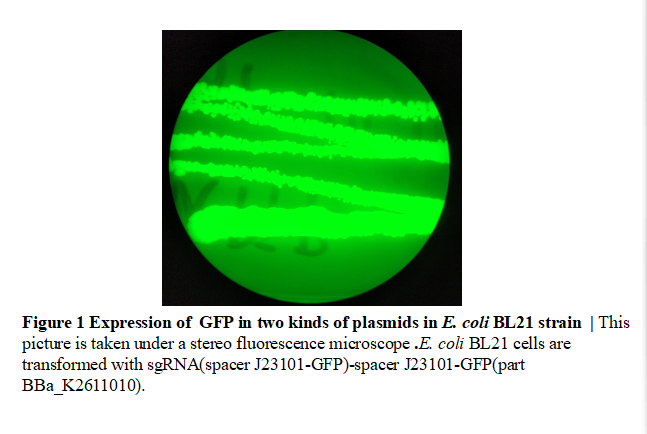
| − | We added a spacer sequence and a NGG site closely before the promoter ( | + | We added a spacer sequence and a NGG site closely before the promoter (J23101). This spacer sequence can be recognized by the sgRNA we designed to direct dCas9. Once dCas9 binds with the spacer sequence, the expression of GFP will be repressed.We have submitted the sgRNA part (BBa_K2611000) and spacer-GFP part(BBa_K2611001). In this composite part, we linked them together.We observed positive colonies transformed with sgRNA(spacer J23101-GFP)-spacer J23101-GFP(part BBa_K2611010) under a stereo fluorescence microscope.Green fluorescence was observed.(Fig.1) |
| − | ). This spacer sequence can be recognized by the sgRNA we designed to direct dCas9. Once dCas9 binds with the spacer sequence, the expression of GFP will be repressed.We have submitted the sgRNA part (BBa_K2611000) and spacer-GFP part(BBa_K2611001). In this composite part, we linked them together.We observed positive colonies transformed with sgRNA(spacer J23101-GFP)-spacer J23101-GFP(part BBa_K2611010) under a stereo fluorescence microscope.Green fluorescence was observed.(Fig.1) | + | |
[[File:Long description.png]] | [[File:Long description.png]] | ||
Revision as of 07:52, 17 October 2018
sgRNA(spacer J23101-GFP)-spacer J23101-GFP
We added a spacer sequence and a NGG site closely before the promoter (J23101). This spacer sequence can be recognized by the sgRNA we designed to direct dCas9. Once dCas9 binds with the spacer sequence, the expression of GFP will be repressed.We have submitted the sgRNA part (BBa_K2611000) and spacer-GFP part(BBa_K2611001). In this composite part, we linked them together.We observed positive colonies transformed with sgRNA(spacer J23101-GFP)-spacer J23101-GFP(part BBa_K2611010) under a stereo fluorescence microscope.Green fluorescence was observed.(Fig.1)
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 226
Illegal NheI site found at 249 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 924