Difference between revisions of "Part:BBa K2617017"
(→Vector design for the validation of expression system) |
|||
| Line 14: | Line 14: | ||
==Characterization== | ==Characterization== | ||
| − | We have improved the previous part [https://parts.igem.org/Part:BBa_J23100 BBa_J23100] by adding RBS and pelB-5D to make it can be used for extracelluar expression. The combination of promoter, RBS and signal peptide makes it convenient to use as a | + | We have improved the previous part [https://parts.igem.org/Part:BBa_J23100 BBa_J23100] by adding RBS and pelB-5D to make it can be used for extracelluar expression. The combination of promoter, RBS and signal peptide makes it convenient to use as a Biobrick and gives it the function of extracellular expression. |
| − | |||
For the function determination, we chose the PETase as our reporter protein because its activity can be easily detected. Thus we constructed two plasmids with and without pelB-5D respectively to compare the function of our improved part and previous part. | For the function determination, we chose the PETase as our reporter protein because its activity can be easily detected. Thus we constructed two plasmids with and without pelB-5D respectively to compare the function of our improved part and previous part. | ||
| Line 41: | Line 40: | ||
===Experiment Result=== | ===Experiment Result=== | ||
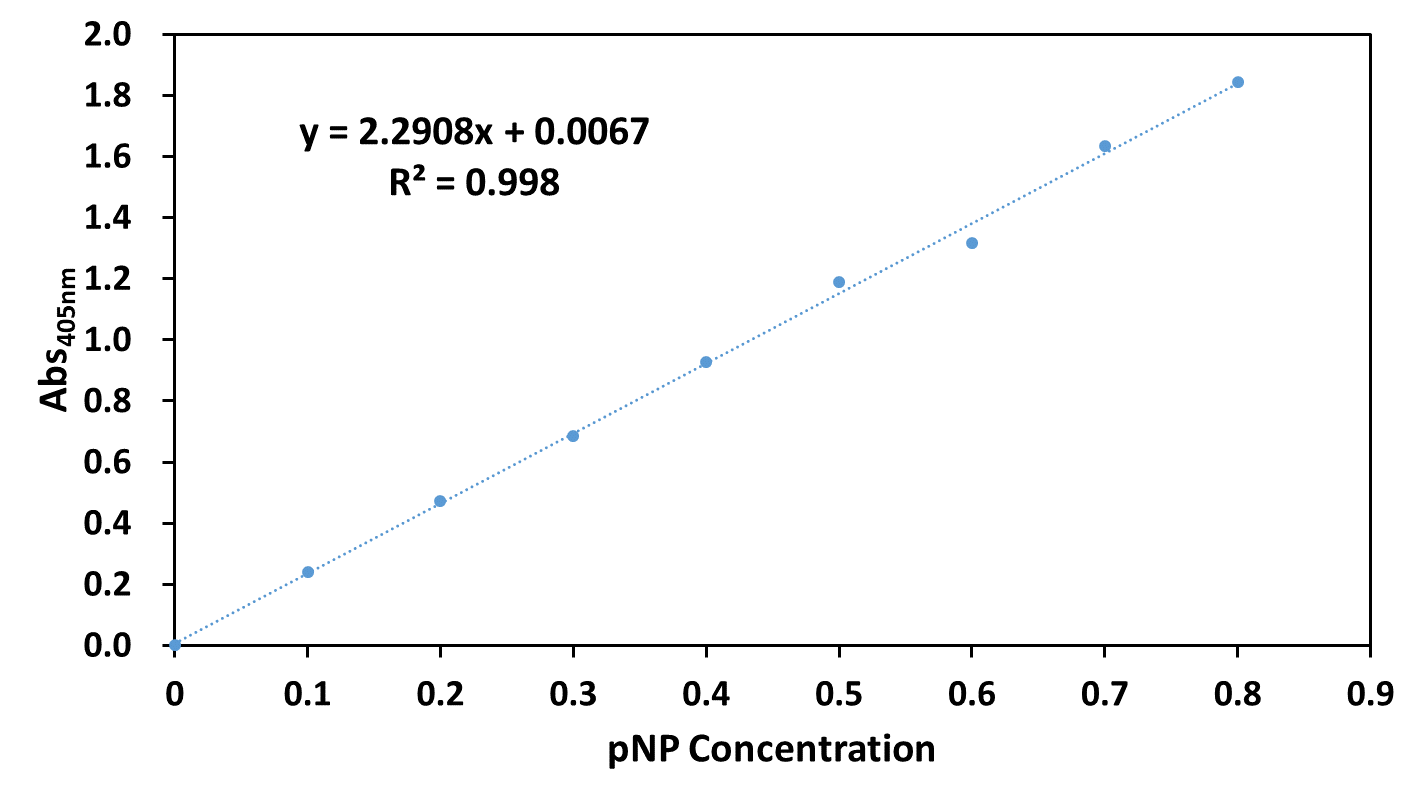
For quantitative assay, a standard curve of pNP ranging from 0-0.8 mM in 100 mM of phosphate buffer (pH 7.4) was measured. | For quantitative assay, a standard curve of pNP ranging from 0-0.8 mM in 100 mM of phosphate buffer (pH 7.4) was measured. | ||
| − | + | [[File:T--UESTC-China--improvet1.png|500px|center|'''Fig. 1 The standard curve of pNP.]] | |
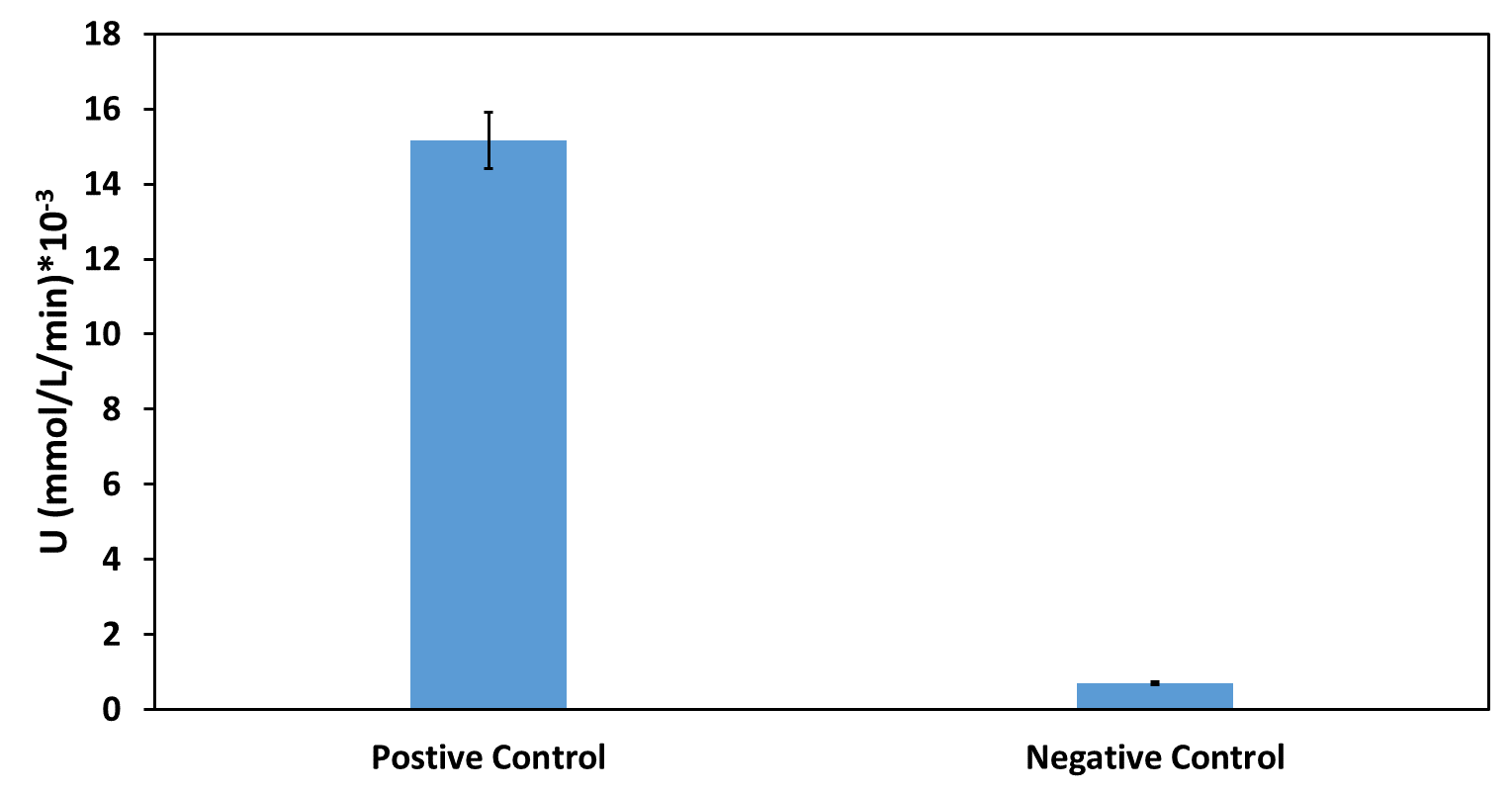
| − | + | The activity of the extracelluar fractions was detected by the method of PETase. And the results were as followed. | |
| − | + | [[File:T--UESTC-China--improvet2.png|500px|center|'''Fig. 2 The activity for extracellular fractions of Positive Control and Negative Control.]] | |
| − | + | Based on our functional determination test results, the improvement that we did has succeeded. As you could see, PETase that carrying our improved part (Fig. 2) has much higher activity level than the PETase that carrying promoter BBa_J23100 but without pelB-5D. From our experiment result, we could conclude that our improved part indeed gives the promoter BBa_J23100 the function of extracellular expression. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 13:21, 16 October 2018
J23100-RBS-pelB-5D
A combination system of promoter, ribosome binding site and signal peptide for enhancing protein expression(pelB)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 115
- 1000COMPATIBLE WITH RFC[1000]
Characterization
We have improved the previous part BBa_J23100 by adding RBS and pelB-5D to make it can be used for extracelluar expression. The combination of promoter, RBS and signal peptide makes it convenient to use as a Biobrick and gives it the function of extracellular expression.
For the function determination, we chose the PETase as our reporter protein because its activity can be easily detected. Thus we constructed two plasmids with and without pelB-5D respectively to compare the function of our improved part and previous part.
| No. | Vector | E.coli resistance | Vector map | Description |
|---|---|---|---|---|
Experiment Result
For quantitative assay, a standard curve of pNP ranging from 0-0.8 mM in 100 mM of phosphate buffer (pH 7.4) was measured.
The activity of the extracelluar fractions was detected by the method of PETase. And the results were as followed.
Based on our functional determination test results, the improvement that we did has succeeded. As you could see, PETase that carrying our improved part (Fig. 2) has much higher activity level than the PETase that carrying promoter BBa_J23100 but without pelB-5D. From our experiment result, we could conclude that our improved part indeed gives the promoter BBa_J23100 the function of extracellular expression.