Difference between revisions of "Reading Gel Images"
(New page: As part of the 2008 quality control project, each part was digested with Eco RI and Ban I restriction enzymes. We then ran all of these digested parts out on gels in order to determine if...) |
|||
| Line 1: | Line 1: | ||
As part of the 2008 quality control project, each part was digested with Eco RI and Ban I restriction enzymes. We then ran all of these digested parts out on gels in order to determine if the insertions and plasmids were correct. | As part of the 2008 quality control project, each part was digested with Eco RI and Ban I restriction enzymes. We then ran all of these digested parts out on gels in order to determine if the insertions and plasmids were correct. | ||
| − | + | First check to see the which lane your part of interest is loaded into. Due to the way the gels were loaded they are not in order by the wells on the plate. For example, if you are looking at part I718014, which is located in well 1F of plate 1007, you would go to lane 11 in gel 1007 1-6. | |
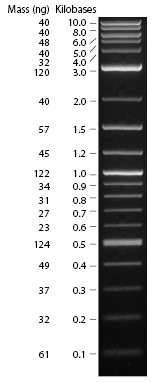
| − | + | Find the lane containing the part and measure the size of the bands by comparing them to the ladder. The ladder was loaded into the lanes on the ends of the gel. See image for the size of bands. | |
[[Image:N3200_fig1_v1_000036.gif]] | [[Image:N3200_fig1_v1_000036.gif]] | ||
| − | + | Compare the size of the bands from the gel to the size of the insert and plasmid for the part. For example, for part I718014 the insert is 919 base pairs and the plasmid is 2058 base pairs. The gel shows bands a little above 900 bp and a little above 2000 bp, so we can confirm this part. Had one or more of the bands been in the wrong place, then the insertion, plasmid, or both would be marked as bad. | |
| − | + | If more than two bands are present than the part was marked as questionable or bad, depending upon the lengths of the bands. | |
| − | + | If no bands were present then the part was marked as bad. | |
| − | + | The quality or relative concentration of DNA was also measured. If the band appeared to be faint, then it has a low concentration of DNA. If the band is dense and bright then it is considered to have a high concentration of DNA. | |
Latest revision as of 19:44, 7 July 2008
As part of the 2008 quality control project, each part was digested with Eco RI and Ban I restriction enzymes. We then ran all of these digested parts out on gels in order to determine if the insertions and plasmids were correct.
First check to see the which lane your part of interest is loaded into. Due to the way the gels were loaded they are not in order by the wells on the plate. For example, if you are looking at part I718014, which is located in well 1F of plate 1007, you would go to lane 11 in gel 1007 1-6.
Find the lane containing the part and measure the size of the bands by comparing them to the ladder. The ladder was loaded into the lanes on the ends of the gel. See image for the size of bands.
Compare the size of the bands from the gel to the size of the insert and plasmid for the part. For example, for part I718014 the insert is 919 base pairs and the plasmid is 2058 base pairs. The gel shows bands a little above 900 bp and a little above 2000 bp, so we can confirm this part. Had one or more of the bands been in the wrong place, then the insertion, plasmid, or both would be marked as bad.
If more than two bands are present than the part was marked as questionable or bad, depending upon the lengths of the bands.
If no bands were present then the part was marked as bad.
The quality or relative concentration of DNA was also measured. If the band appeared to be faint, then it has a low concentration of DNA. If the band is dense and bright then it is considered to have a high concentration of DNA.