Difference between revisions of "Part:BBa K2549006"
(→Biology) |
(→Biology) |
||
| Line 12: | Line 12: | ||
===Biology=== | ===Biology=== | ||
=====Significance of Notch signaling ===== | =====Significance of Notch signaling ===== | ||
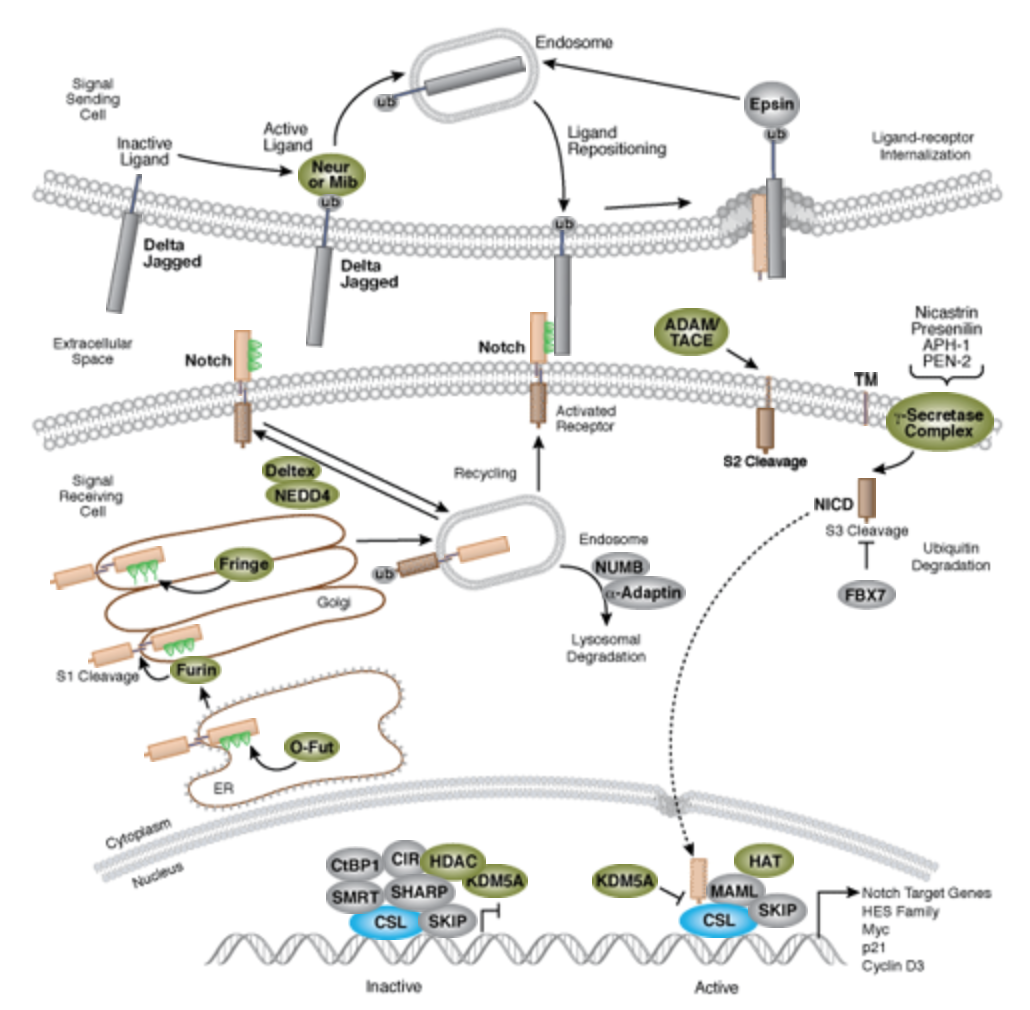
| + | The Notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms. Mammals possess four different notch receptors, referred to as NOTCH1 to NOTCH4. The notch receptor is a single-pass transmembrane receptor protein<ref>https://en.wikipedia.org/wiki/Notch_signaling_pathway</ref>. | ||
| + | [[[[File:Notch-signal.png|none|400px|thumb|''Notch signaling is an evolutionarily conserved pathway in multicellular organisms that regulates cell-fate determination during development and maintains adult tissue homeostasis. The Notch pathway mediates juxtacrine cellular signaling wherein both the signal sending and receiving cells are affected through ligand-receptor crosstalk by which an array of cell fate decisions in neuronal, cardiac, immune, and endocrine development are regulated''<ref>https://www.cellsignal.com/contents/science-cst-pathways-stem-cell-markers/notch-signaling-interactive-pathway/pathways-notch</ref>.]] | ||
| + | |||
| + | Dr. Hans Widlund (Brigham and Women’s Hospital, Harvard Medical School) once contributed a list of original articles worth reading. There are significant amount of Notch research articles and reviews in PubMed<ref>https://www.ncbi.nlm.nih.gov/pubmed/?term=notch</ref> as well. | ||
| + | * Ables JL, Breunig JJ, Eisch AJ, Rakic P (2011) Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12(5), 269–83. | ||
| + | * Andersson ER, Lendahl U (2014) Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov 13(5), 357–78. | ||
| + | * Aster JC, Blacklow SC, Pear WS (2011) Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223(2), 262–73. | ||
| + | * Bai G, Pfaff SL (2011) Protease regulation: the Yin and Yang of neural development and disease. Neuron 72(1), 9–21. | ||
| + | * de la Pompa JL, Epstein JA (2012) Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22(2), 244–54. | ||
| + | * Ntziachristos P, Lim JS, Sage J, Aifantis I (2014) From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25(3), 318–34. | ||
| + | * Ranganathan P, Weaver KL, Capobianco AJ (2011) Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11(5), 338–51. | ||
| + | * Weinmaster G, Fischer JA (2011) Notch ligand ubiquitylation: what is it good for? Dev. Cell 21(1), 134–44. | ||
| + | * Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ (2010) Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 28, 343–65. | ||
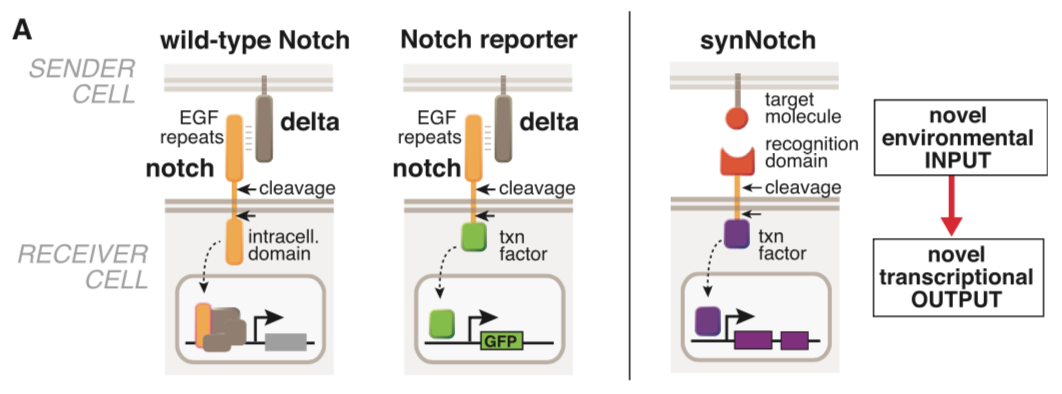
===== synNotch with α-CD19 against CD19 antigen works extremely well in Morsut L et al 2016===== | ===== synNotch with α-CD19 against CD19 antigen works extremely well in Morsut L et al 2016===== | ||
| Line 21: | Line 34: | ||
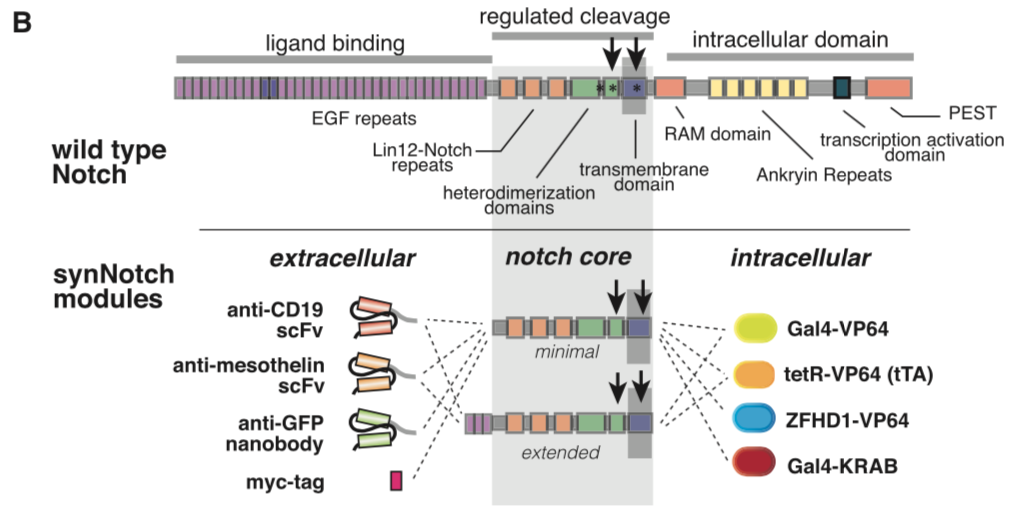
[[File:synNotch2.png|none|400px|thumb|Morsut L at al stated: ''Modularity of the synNotch platform: the input and output domains from Notch can be swapped with diverse domains. On the extracellular side, diverse recognition domains can be used (antibody based, or peptide tags are shown), and on the intracellular side, diverse effector can be used (transcriptional activators with different DBDs are shown, as well as a transcriptional repressor).'']] | [[File:synNotch2.png|none|400px|thumb|Morsut L at al stated: ''Modularity of the synNotch platform: the input and output domains from Notch can be swapped with diverse domains. On the extracellular side, diverse recognition domains can be used (antibody based, or peptide tags are shown), and on the intracellular side, diverse effector can be used (transcriptional activators with different DBDs are shown, as well as a transcriptional repressor).'']] | ||
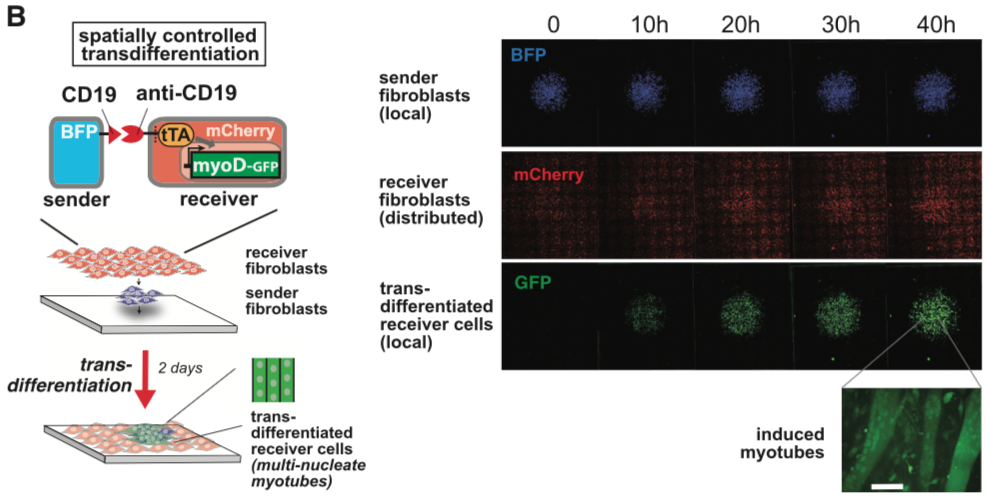
| − | [[File:synNotch3.png|none| | + | [[File:synNotch3.png|none|250px|thumb|Morsut L at al stated: ''SynNotch receptors can be used to detect endogenous disease antigens and induce the expression of a reporter gene. Mouse fibroblasts (L929 line) expressing anti-CD19/tTA synNotch are cultivated with K562 sender cells expressing Delta, CD19, or CD19 in the presence of the gamma-secretase inhibitor DAPT. Fluorescence-activated cell sorting (FACS) plots of the resulting GFP reporter intensity in receiver cells are shown. Inset shows an image of MDCK cells expressing the anti-CD19/GFP synNotch, when co-cultivated for 24 hr with MDCK sender cells expressing CD19 (constitutively labeled with tagBFP). Only receiver cells in contact with (blue) sender cells activate the reporter and turn green.'']] |
| − | [[File:synNotch4.png|none| | + | [[File:synNotch4.png|none|200px|thumb|Morsut L at al stated: ''Stimulation of mouse fibroblasts expressing anti-GFP synNotch with ligands in different formats. Anti-GFP synNotch receiver cells are stimulated for 1 hr with GFP either in soluble form, presented on a K562 sender-cell, or cis-presented on the receiver cell itself. The receiver cells show activation only when the ligand is present on an opposing surface and if they do not express the ligand in cis. The FACS data are recorded at 24 hr after the beginning of stimulation. FACS histograms include at least 10,000 cells for each condition.'']] |
[[File:synNotch5.png|none|400px|thumb|Morsut L at al stated: ''Mouse fibroblasts (L929 line) with anti-CD19 synNotch with a transcriptional repressor intracellular domain (Gal4-KRAB) are co-cultivated with K562 sender cells. The receiver cells constitutively express GFP downstream of a SV40/UAS combined promoter. FACS plot of receiver cells is shown, in presence of K562 sender cells with or without CD19 expression, as indicated in figure.'']] | [[File:synNotch5.png|none|400px|thumb|Morsut L at al stated: ''Mouse fibroblasts (L929 line) with anti-CD19 synNotch with a transcriptional repressor intracellular domain (Gal4-KRAB) are co-cultivated with K562 sender cells. The receiver cells constitutively express GFP downstream of a SV40/UAS combined promoter. FACS plot of receiver cells is shown, in presence of K562 sender cells with or without CD19 expression, as indicated in figure.'']] | ||
Revision as of 09:09, 7 October 2018
mouse Notch1 core
This part is the mouse Notch1 minimal regulatory region[1]. It was used as the transmembrane core domain of the SynNotch. However, original reported design has a high background activation in transfected 293T cells. We have put a lot of effect in optimizing this critical transmembrane domain for Notch receptor.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 592
Biology
Significance of Notch signaling
The Notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms. Mammals possess four different notch receptors, referred to as NOTCH1 to NOTCH4. The notch receptor is a single-pass transmembrane receptor protein[2].
[[
Dr. Hans Widlund (Brigham and Women’s Hospital, Harvard Medical School) once contributed a list of original articles worth reading. There are significant amount of Notch research articles and reviews in PubMed[4] as well.
- Ables JL, Breunig JJ, Eisch AJ, Rakic P (2011) Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12(5), 269–83.
- Andersson ER, Lendahl U (2014) Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov 13(5), 357–78.
- Aster JC, Blacklow SC, Pear WS (2011) Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223(2), 262–73.
- Bai G, Pfaff SL (2011) Protease regulation: the Yin and Yang of neural development and disease. Neuron 72(1), 9–21.
- de la Pompa JL, Epstein JA (2012) Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22(2), 244–54.
- Ntziachristos P, Lim JS, Sage J, Aifantis I (2014) From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25(3), 318–34.
- Ranganathan P, Weaver KL, Capobianco AJ (2011) Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11(5), 338–51.
- Weinmaster G, Fischer JA (2011) Notch ligand ubiquitylation: what is it good for? Dev. Cell 21(1), 134–44.
- Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ (2010) Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 28, 343–65.
synNotch with α-CD19 against CD19 antigen works extremely well in Morsut L et al 2016
Please refer the original article for more details.






References
- ↑ Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Morsut L, Roybal KT, Xiong X, ..., Thomson M, Lim WA. Cell, 2016 Feb;164(4):780-91 PMID: 26830878; DOI: 10.1016/j.cell.2016.01.012
- ↑ https://en.wikipedia.org/wiki/Notch_signaling_pathway
- ↑ https://www.cellsignal.com/contents/science-cst-pathways-stem-cell-markers/notch-signaling-interactive-pathway/pathways-notch
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/?term=notch

