Difference between revisions of "Part:BBa K2888009"
Yinchizhou18 (Talk | contribs) |
|||
| Line 5: | Line 5: | ||
This mlrA gene is a part of the microcystin degradation system discovered in Sphingopyxis sp. C-1. In our experiment, we intend to deal with Microcystin-LR, the most common type of microcystin toxin produced in cyanobacteria. The mlrA gene is responsible for hydrolytic cleavage and linearization of cyclic structure of MCLR. | This mlrA gene is a part of the microcystin degradation system discovered in Sphingopyxis sp. C-1. In our experiment, we intend to deal with Microcystin-LR, the most common type of microcystin toxin produced in cyanobacteria. The mlrA gene is responsible for hydrolytic cleavage and linearization of cyclic structure of MCLR. | ||
| + | |||
| + | |||
| + | ===Introduction=== | ||
| + | Microcystin(MC) degradation system was investigated in the MC-degrading bacteria species, Sphingopyxis sp. C-1, in previous experiment(Shimizu 2011).Among 10-more species of such bacteria, our group intends to research on the strand Sphingomonas sp, which possess a MC-degrading pathway that involves 3 genes: mrl A mrl B mrl C that works in a sequential chain reaction(Shimizu 2011) and an additional gene, mrl D, whose product is speculated to aid the transport of MCLR or its degradation process(Bourne, 2001). | ||
| + | |||
| + | https://www.omicsonline.org/publication-images/environmental-analytical-toxicology-General-chemical-8-556-g001.png | ||
| + | |||
| + | ===Our Design=== | ||
| + | |||
| + | Based on previous experiment, we add constitutive promoter, RBS, and double terminator to mlrA gene | ||
| + | |||
| + | |||
| + | ===Our Experience=== | ||
| + | We successfully infused mlrA gene with pSB1C3 backbone and transformed it into DH5 alpha E.coli in order to get plasmids. Then we transform extracted plasmids into BL21 E.coli in order to culture abundant E.coli and expressed proteins which contain our target gene. Although, the purification of the protein is sill in process due to limited time. | ||
| + | |||
| + | <html> | ||
| + | <h4>Protocol</h4> | ||
| + | <p> 1. Amplify target gene and pSB1C3 backbone </p> | ||
| + | <p>2. Gel electrophoresis to verify the existence of amplified gene</p> | ||
| + | <p>2. Infusion of backbone and target gene</p> | ||
| + | <p>3. Transformation and single colony selection</p> | ||
| + | <p>4. Inoculation in liquid LB culture</p> | ||
| + | <p>5. Bacteria PCR to verify the existence of amplified gene</p> | ||
| + | <p>6. Extraction of plasmids and transformation into BL-21 E.coli</p> | ||
| + | <p>7. Growth of E.coli</p> | ||
| + | <p>8. Protein Purification</p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | ===Reference=== | ||
| + | Shimizu, K. et al. How microcystin-degrading bacteria express microcystin degradation activity. Lakes & Reservoirs: Research & Management 16, 169–178 (2011). | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Revision as of 20:42, 17 October 2018
fused mlrA gene
This mlrA gene is a part of the microcystin degradation system discovered in Sphingopyxis sp. C-1. In our experiment, we intend to deal with Microcystin-LR, the most common type of microcystin toxin produced in cyanobacteria. The mlrA gene is responsible for hydrolytic cleavage and linearization of cyclic structure of MCLR.
Introduction
Microcystin(MC) degradation system was investigated in the MC-degrading bacteria species, Sphingopyxis sp. C-1, in previous experiment(Shimizu 2011).Among 10-more species of such bacteria, our group intends to research on the strand Sphingomonas sp, which possess a MC-degrading pathway that involves 3 genes: mrl A mrl B mrl C that works in a sequential chain reaction(Shimizu 2011) and an additional gene, mrl D, whose product is speculated to aid the transport of MCLR or its degradation process(Bourne, 2001).
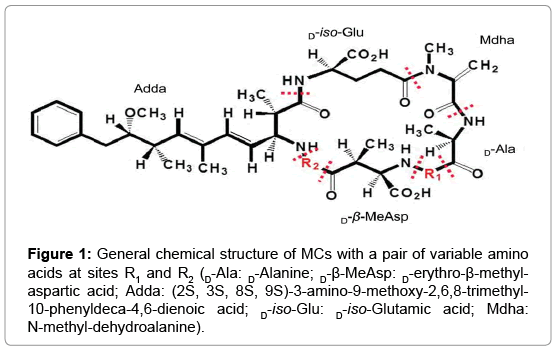
Our Design
Based on previous experiment, we add constitutive promoter, RBS, and double terminator to mlrA gene
Our Experience
We successfully infused mlrA gene with pSB1C3 backbone and transformed it into DH5 alpha E.coli in order to get plasmids. Then we transform extracted plasmids into BL21 E.coli in order to culture abundant E.coli and expressed proteins which contain our target gene. Although, the purification of the protein is sill in process due to limited time.
Protocol
1. Amplify target gene and pSB1C3 backbone
2. Gel electrophoresis to verify the existence of amplified gene
2. Infusion of backbone and target gene
3. Transformation and single colony selection
4. Inoculation in liquid LB culture
5. Bacteria PCR to verify the existence of amplified gene
6. Extraction of plasmids and transformation into BL-21 E.coli
7. Growth of E.coli
8. Protein Purification
Reference
Shimizu, K. et al. How microcystin-degrading bacteria express microcystin degradation activity. Lakes & Reservoirs: Research & Management 16, 169–178 (2011). Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 770
Illegal AgeI site found at 305 - 1000COMPATIBLE WITH RFC[1000]
