Difference between revisions of "Part:BBa I13521"
Laurent CHEN (Talk | contribs) (→Pictures) |
|||
| Line 20: | Line 20: | ||
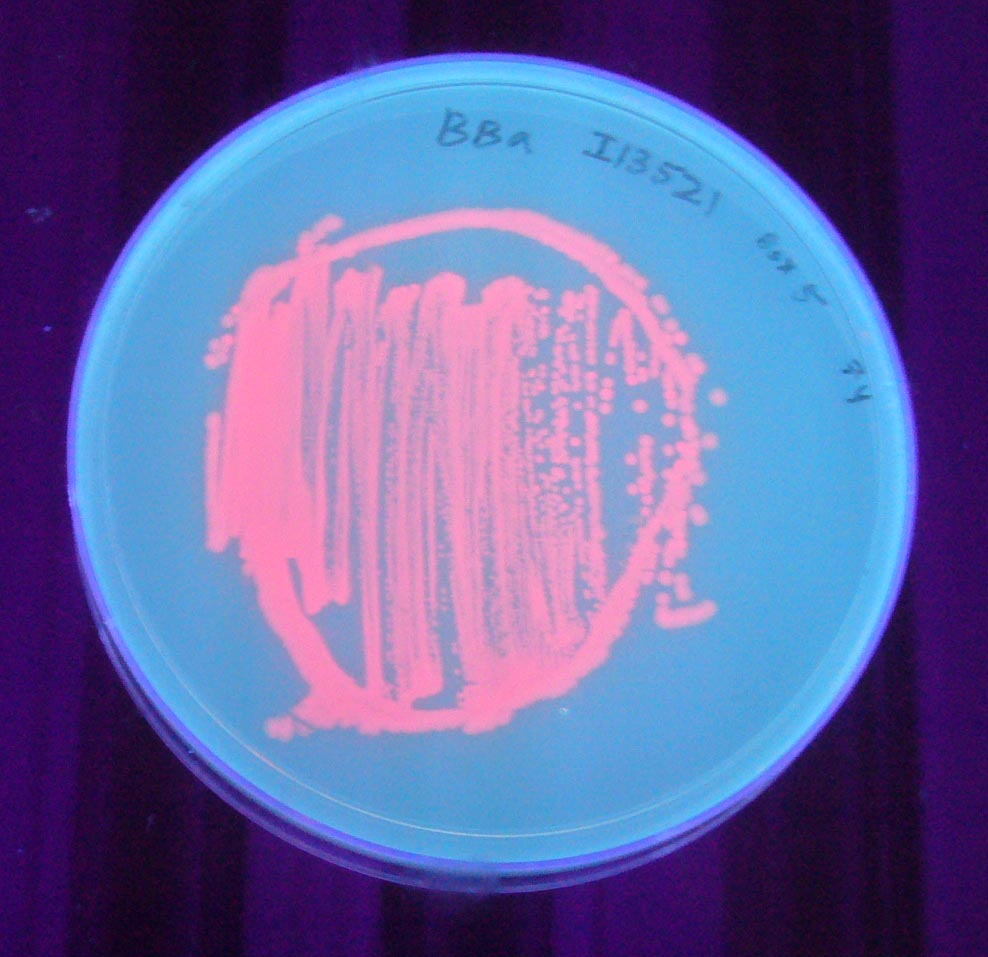
Image:I13521 - UV 254nm.jpg|BBa_I13521 visualized under 254nm wavelength UV lightbox | Image:I13521 - UV 254nm.jpg|BBa_I13521 visualized under 254nm wavelength UV lightbox | ||
</gallery> | </gallery> | ||
| + | |||
| + | <h2> Part characterization by iGEM Sorbonne Université 2019 </h2> | ||
| + | <br> | ||
| + | Day 1: <br> | ||
| + | The DNA corresponding to this part is amplified and extracted from liquid cultures of bacteria thanks to the Promega extraction kit.<br> | ||
| + | Control digests by Eco RI and Pst I were carried out in order to verify the presence of the part of 923 bp. DNA fragments were analyzed on a 1.2% agarose gel. The 8 clones tested have the correct BBa_I13521.<br> | ||
| + | <center> </center> | ||
| + | <b><i>Controls digestions of DNA from the clones 1 and 2.<b><i><br> | ||
| + | <center> </center> | ||
| + | <b><i>Controls digestions of DNA from the clones 3 to 8.<b><i><br> | ||
| + | |||
| + | The plasmid DNAs containing part 1 to 8 are used to transform competent DH5 α bacteria which are then plated on LB medium + Chloramphenicol 34 μg/mL and incubated overnight at 37°C.<br><br> | ||
| + | |||
| + | Day 2:<br> | ||
| + | A clone resulting from each transformation is subcultured in liquid culture of 5 mL of LB medium + Chloramphenicol 20 μg/mL. These cultures are incubated overnight at 37°C with 200 rpm shaking.<br><br> | ||
| + | |||
| + | Day 3: <br> | ||
| + | The 8 liquid cultures are diluted to an OD at 600 nm of 0.1 in LB medium + Chloramphenicol 20 μg/mL.<br> | ||
| + | The liquid cultures are then placed at 37°C. with stirring at 200 rpm until an OD600 of 0.4 is reached.<br> | ||
| + | Each of the 8 liquid cultures is divided into 3, corresponding to 3 different concentrations of tetracycline: 0 ng/mL - 100 ng/mL – 10 000 ng/mL.<br> | ||
| + | A time course experiments is realized, measuring the OD600 and mRFP fluorescence (535 nm (excitation) / 620 nm (emission) of 100µL of each culture every 30min.<br> | ||
| + | As expected, the more tetracycline there was, the slowly the OD 600 increased, meaning that even at this low concentration, the tetracycline is toxic for bacteria. mRFP fluorescence was too low to overpass background signal (data not shown).<br> | ||
| + | To check if bacteria were expressing the mRFP, we observed the cells after 6h incubation at 37°C using an epi-fluorescence microscope. Doing so, we were able to detect mRFP signal, the bacteria being red. Therefore, we quantified the mRFP fluorescence signal for the bacteria transformed with the BBa_I13521 part.<br> | ||
| + | |||
| + | <center>https://static.igem.org/mediawiki/parts/5/52/T--Sorbonne_U_Paris--Contribution_Final.png</center> | ||
| + | |||
| + | We observed no significative variation of the mRFP signal depending on the tetracycline concentration.<br> | ||
| + | The BBa_I13521 part is described as “Constitutive on mRFP (positive control)” but the subpart description is not coherent with this description, as it is stated that this reporter construct has a ptetR promoter (regulated by tetracycline, thus not constitutive). The two description being incoherent, we wanted to identify the correct one.<b> Our data are in favor of a constitutive expression of the mRFP, not affected by the addition of tetracycline.</b> | ||
<h2>The followings were edited by iGEM17_SCU-China</h2> | <h2>The followings were edited by iGEM17_SCU-China</h2> | ||
Revision as of 15:43, 17 October 2019
Ptet mRFP
Constitutive on mRFP (positive control)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 635
Illegal AgeI site found at 747 - 1000COMPATIBLE WITH RFC[1000]
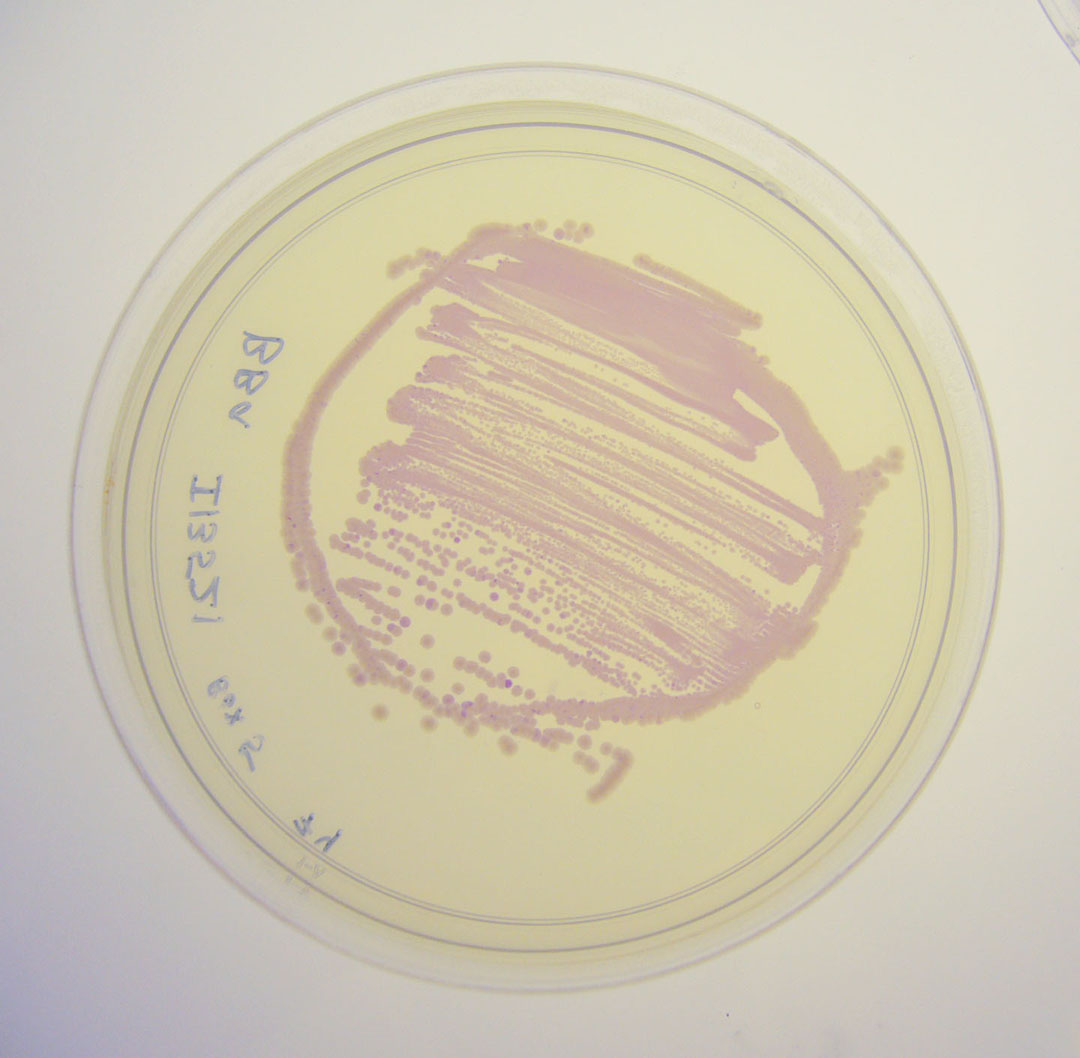
Pictures
Part characterization by iGEM Sorbonne Université 2019
Day 1:
The DNA corresponding to this part is amplified and extracted from liquid cultures of bacteria thanks to the Promega extraction kit.
Control digests by Eco RI and Pst I were carried out in order to verify the presence of the part of 923 bp. DNA fragments were analyzed on a 1.2% agarose gel. The 8 clones tested have the correct BBa_I13521.
Controls digestions of DNA from the clones 1 and 2.<b><i>
<b><i>Controls digestions of DNA from the clones 3 to 8.<b><i>
The plasmid DNAs containing part 1 to 8 are used to transform competent DH5 α bacteria which are then plated on LB medium + Chloramphenicol 34 μg/mL and incubated overnight at 37°C.
Day 2:
A clone resulting from each transformation is subcultured in liquid culture of 5 mL of LB medium + Chloramphenicol 20 μg/mL. These cultures are incubated overnight at 37°C with 200 rpm shaking.
Day 3:
The 8 liquid cultures are diluted to an OD at 600 nm of 0.1 in LB medium + Chloramphenicol 20 μg/mL.
The liquid cultures are then placed at 37°C. with stirring at 200 rpm until an OD600 of 0.4 is reached.
Each of the 8 liquid cultures is divided into 3, corresponding to 3 different concentrations of tetracycline: 0 ng/mL - 100 ng/mL – 10 000 ng/mL.
A time course experiments is realized, measuring the OD600 and mRFP fluorescence (535 nm (excitation) / 620 nm (emission) of 100µL of each culture every 30min.
As expected, the more tetracycline there was, the slowly the OD 600 increased, meaning that even at this low concentration, the tetracycline is toxic for bacteria. mRFP fluorescence was too low to overpass background signal (data not shown).
To check if bacteria were expressing the mRFP, we observed the cells after 6h incubation at 37°C using an epi-fluorescence microscope. Doing so, we were able to detect mRFP signal, the bacteria being red. Therefore, we quantified the mRFP fluorescence signal for the bacteria transformed with the BBa_I13521 part.

We observed no significative variation of the mRFP signal depending on the tetracycline concentration.
The BBa_I13521 part is described as “Constitutive on mRFP (positive control)” but the subpart description is not coherent with this description, as it is stated that this reporter construct has a ptetR promoter (regulated by tetracycline, thus not constitutive). The two description being incoherent, we wanted to identify the correct one.<b> Our data are in favor of a constitutive expression of the mRFP, not affected by the addition of tetracycline.</b>
The followings were edited by iGEM17_SCU-China
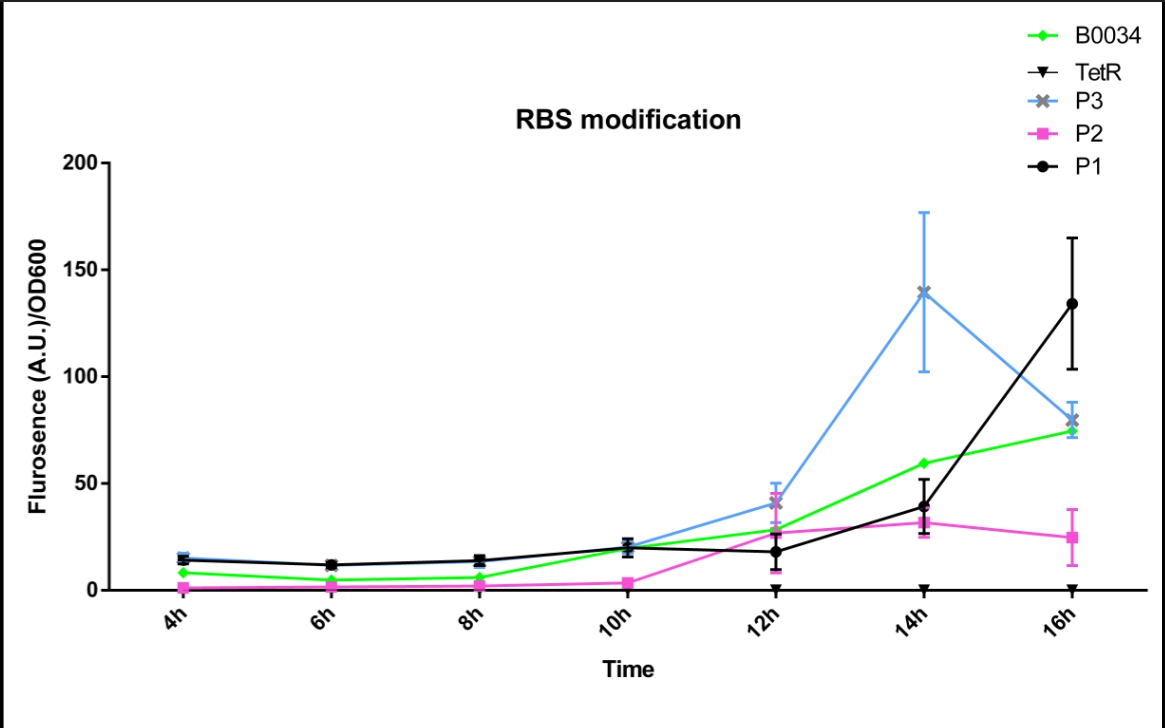
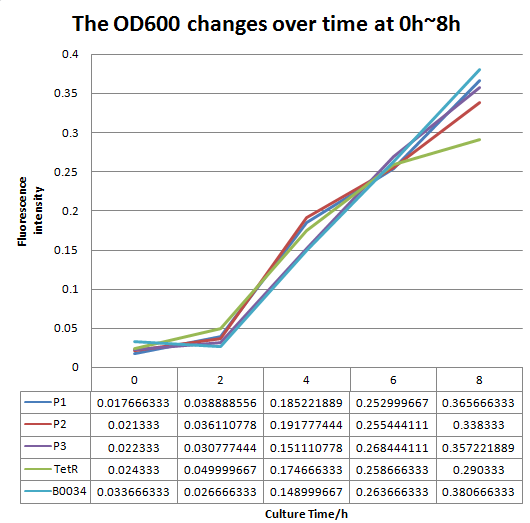
Improvement
We altered the RBS region and constructed new parts that has higher translation rate, so that the expression of this part, BBa_I13521, could increase, which may benefit the properties of BBa_I13521 as a reporter device with higher sensitivity. To test the performance, we measure the fluorescence intensity and OD600 changes of the bacteria liquid over time within 16hours after inoculated. The result showed that we indeed improved the Part:BBa_I13521 and got a new part, BBa_K2276010, containing a stronger RBS. Fluorescence quenching may explain the drop of the fluorescence intensity of P3 strain at 16h because the concentration of fluorescent protein was too high.