Difference between revisions of "Part:BBa K2457001"
Giomaklouf (Talk | contribs) (→Characterization) |
|||
| Line 28: | Line 28: | ||
===Characterization=== | ===Characterization=== | ||
| + | <p>BBa_K2457001 was used by Amazonas_Brazil team in CROUT experiments and CRIN experiments.</p> | ||
| + | <p>CROUT (CRISPeasy-out method): Designed for knock-out strategy for bacterial genome editing based on CRISPR/Cas9</p> | ||
| + | |||
| + | <p>RESULTADOS DE CROUT AQUI JU</p> | ||
| + | |||
| + | <p>CRIN (CRISPeasy-in method): Designed for knock-in strategy for bacterial genome editing based on CRISPR/Cas9</p> | ||
| + | |||
| + | <p>RESULTADOS DE CRIN AQUI JU</p> | ||
===Sequencing=== | ===Sequencing=== | ||
Revision as of 21:18, 1 November 2017
Standardized Cas9 coding sequence + B0015 terminator
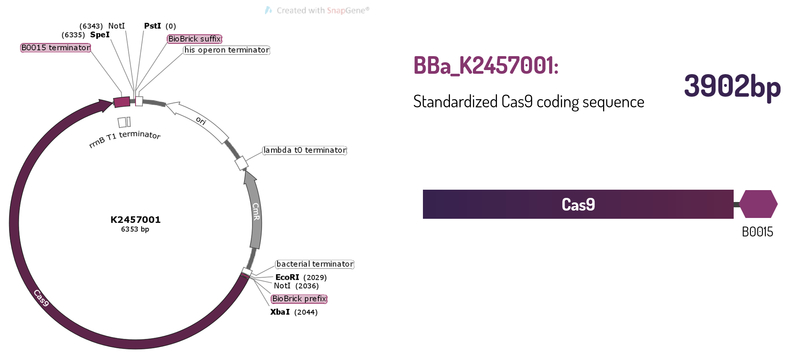
This brick was engineered to be a building block of the standard BioBrick framework for bacterial genome engineering: the ([http://2017.igem.org/Team:Amazonas_Brazil pCRISPeasy]). BBa_K2457001 is composed of the Cas9 coding sequence from Streptococcus pyogenes + BBa_B0015 double terminator. It was rationally designed to be interchangeable AND standardized, providing a BioBrick to attend multiple SynBio purposes. For that matter, the RBS and promoter are easily switchable through 3A Assembly. It’s also standard restriction enzymes safe for all RFCs assemblies.
Figure 1: BBa_K2457001 circuit.
Usage and Biology
In bacteria and archaea, the CRISPR system associated to the Cas proteins is, shortly, a clustered regularly interspaced short palindromic repeats: repeated sequences interspaced by small unrepeated sequences (spacers) resulted from exogenous DNAs segments (protospacers) (Nishimasu et al., 2014). A natural mechanism that promotes immunity through invasors bacteriophage fragments transduction or plasmidial DNA incorporation in prokaryotes CRISPR locus (Church, 2013). The Cas9 (CRISPR-associated protein 9) originated from CRISPR system type II is a tool that has been very explored in genome edition methodologies, it does a sequence-specific cleavage of DNA. Compared to other genomic editing systems, this protein detach itself for the site-specific cleavage programming possibilities through a single guide RNA (sgRNA). Due to its versatility, it is unnecessary to design a new protein to each target sequence, as in the traditional methods, like ZFNs and TALENs. Thus, the genetic edition by Cas9 has been integrating innovative applications.
The crystallography made by Jinek (2014) and Nishumasu et al. (2015) revealed the presence of two lobules in Cas9 protein: the first one is Recognition (REC) dividing itself into three parts, an extensive alpha helix, called bridge helix (60-93 residues), REC1 domain (94-179 and 308-713 residues) and REC2 domain (180-307 residues); the second lobule is from nucleases (NUC), splitting in RuvC in aminoterminal which is divided into three discontinuous segments (RuvC-I to RuvC-III), (1-59, 718-769 and 909-1098 residues), HMN in central part (775-908 residues) and the PAM-interacting domains, which is related to the carboxyterminal (1099-1368 residues).
This part codes for a standardized Cas9-version and a B0015 terminator. It can be applied to genomic editing, especially in bacteria, since Cas9 is central to this tool.
Design
• NheI (GCTAGC), which in the alanine GCT codon (18% de codon usage) was replaced by GCG (32%)
• EcoRI (C-GAATTC), which in arginine CGA codon (30%) was replaced by CGT (24.7%)
• First HindIII (AAGCTT) changed to AAACTT
• Second HindIII (AAGCTT) changed to AGGCTT
• NdeI: CATATG changed to CACATG; CATATG changed to CATACG
• NcoI: CCATGG changed to CCGTGG
• BamHI: GGATCC changed to GGACCC
Characterization
BBa_K2457001 was used by Amazonas_Brazil team in CROUT experiments and CRIN experiments.
CROUT (CRISPeasy-out method): Designed for knock-out strategy for bacterial genome editing based on CRISPR/Cas9
RESULTADOS DE CROUT AQUI JU
CRIN (CRISPeasy-in method): Designed for knock-in strategy for bacterial genome editing based on CRISPR/Cas9
RESULTADOS DE CRIN AQUI JU
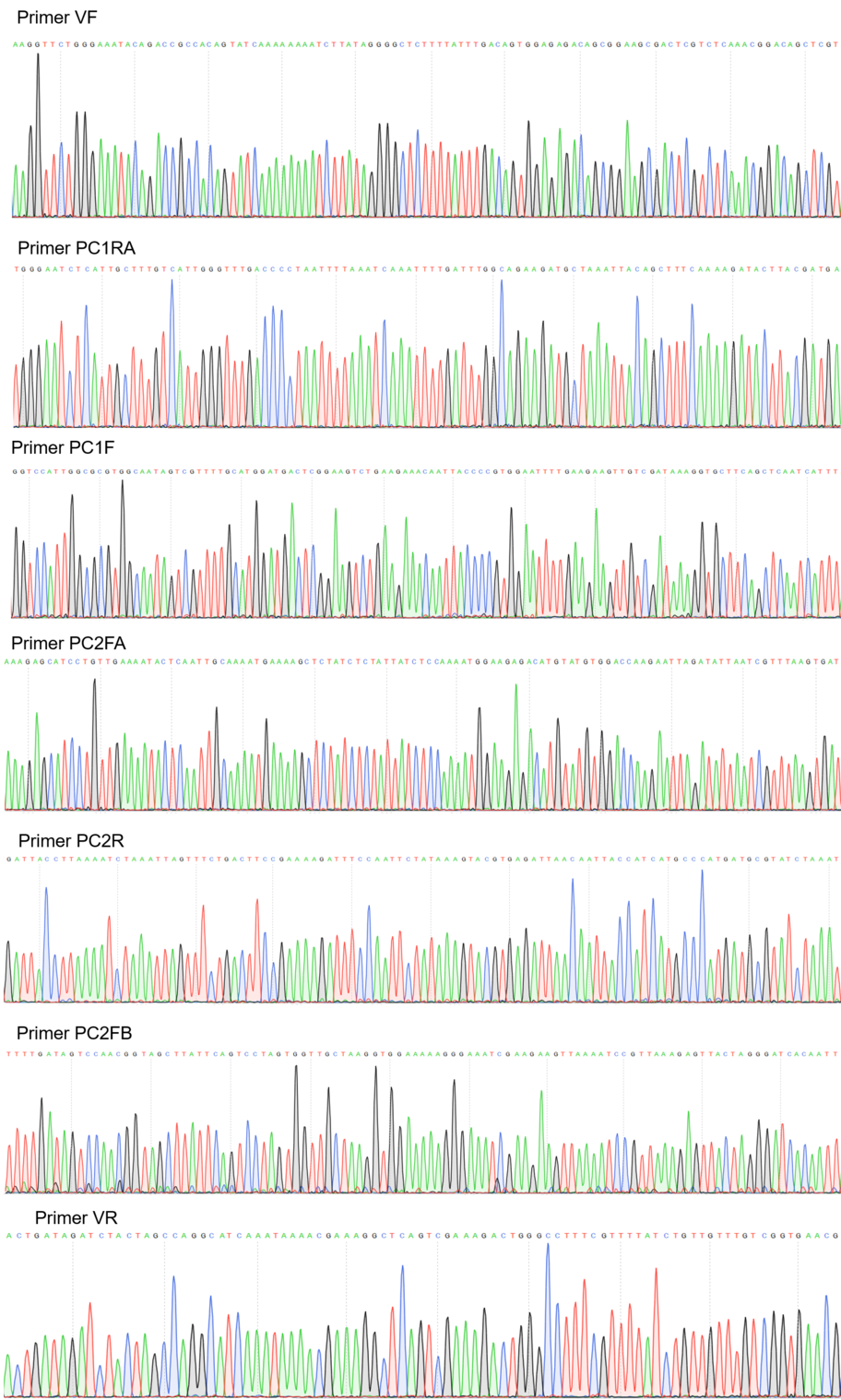
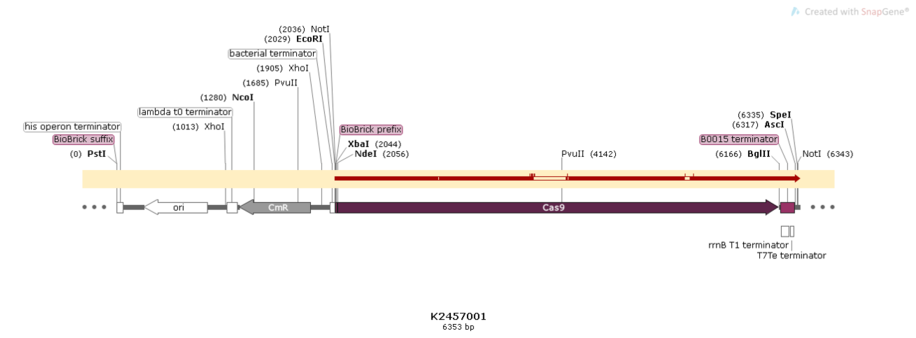
Sequencing
Figure 2:Sequencing electropherogram from BBa_K2457001.
Figure 3:Alignment of the designed sequence and our final construction from BBa_K2457001.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]