Difference between revisions of "Part:BBa K2384011"
| Line 29: | Line 29: | ||
Expression System: <i>Bacillus megaterium</i> | Expression System: <i>Bacillus megaterium</i> | ||
| + | |||
Gene Length:2364 (bp) | Gene Length:2364 (bp) | ||
<br/><br/> | <br/><br/> | ||
| − | + | ||
Show the relative codon used distribution | Show the relative codon used distribution | ||
| − | |||
| − | |||
| − | |||
<table><tr> | <table><tr> | ||
| Line 47: | Line 45: | ||
</tr></table> | </tr></table> | ||
| + | <table><tr> | ||
| + | <th>[[Image:T--FAFU-CHINA-dnamflon.png|thumb|400px|'''Figure 3''':Digested with EcoRI/PstI. ]]</th> | ||
| + | </tr></table> | ||
Revision as of 14:01, 1 November 2017
mf-Lon
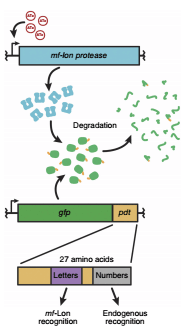
mf-Lon is endogenous Lon protease based on the Gram-positive M.florum tmRNA system.
Usage and Biology
Exogenous control of protein biosynthesis through transcriptional and translational regulation has been well established, but robust and tunable control of protein degradation in bacteria remains elusive.
mf-Lon
Here we present a synthetic degradation system based on the Gram-positive M.florum tmRNA system that does not rely on host degradation systems and function in a wide range of bacteria.
Protein degradation in bacteria occurs in part through the transfer-messenger RNA (tmRNA) system, which uses C-terminal fusion of the ssrA peptide to direct proteins to the endogenous ClpXP and ClpAP proteases for rapid degradation in E. coli. Variants of the
E. coli ssrA tag (ec-ssrA) are commonly used to modify the degradation rate of attached proteins in both bacteria and eukaryotes, but these tags do not provide inducible control of degradation. Recently developed inducible eukaryotic systems rely on degradation machinery not present in bacteria and bacterial systems such as the one developed by Davis et al. require disruption of the endogenous tmRNA system and are therefore not easily transferred to other organisms.
Toggle switch control of targeted essential protein degradation. Inclusion of the mf-Lon–specific pdt#1 tag on the specified essential gene causes mf-Lon–mediated degradation of the essential protein upon circuit activation.
This part is the combination of other two parts(BBa_K2384009) and (BBa_K2384010) , which could work together as a kill switch that uses a transcription-based monostable toggle design to provide rapid and robust target cell killing, dubbed ‘Toggle switch’
This part has made a condon optimization.
Sequence optimization information:
Expression System: Bacillus megaterium
Gene Length:2364 (bp)
Show the relative codon used distribution
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NotI site found at 25
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 2498
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]