Difference between revisions of "Part:BBa I0500:Experience"
(→mCherry) |
|||
| Line 183: | Line 183: | ||
| − | + | = Contribution iGEM TU-Eindhoven 2017 http://2017.igem.org/Team:TU-Eindhoven] = | |
| + | == Method == | ||
| + | We first tested three different temperatures, being 20, 25 and 37 degree Celsius. From our fist basic measurements, we discovered that expression was more efficient at a temperature of 37 degree Celsius and continued with this setting on our incubator. | ||
| + | The next step was consequent adding arabinose to the cultures, as E. coli BL21(DE3) is a strain that can break down arabinose, and for an optimal expression, it is better to have a high enough constant concentration. | ||
| + | We tested an addition of every 60 minutes and every 90 minutes, with varying concentrations of arabinose. | ||
| + | The percentages that are depicted in the figures are the total concentration of arabinose in the whole culture. | ||
== pBAD expression of fluorophore mCherry == | == pBAD expression of fluorophore mCherry == | ||
| − | + | The results of all our tries are depicted in Figure a1 & a2 with the best results in a3. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | The results of all our tries are depicted in Figure a1. | + | |
[[File:T-TU-Eindhoven--pBAD_vector_mCherry_60min.png]] | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_60min.png]] | ||
| Line 200: | Line 200: | ||
[[File:T-TU-Eindhoven--pBAD_vector_mCherry_90min.png]] | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_90min.png]] | ||
| − | ''Figure a2: pBAD expression of | + | ''Figure a2: pBAD expression of mCherry in BL21(DE3) with every 90 minutes an addition of arabinose'' |
[[File:T-TU-Eindhoven--pBAD_vector_mCherry_best_results.png]] | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_best_results.png]] | ||
| Line 206: | Line 206: | ||
''Figure a3: pBAD expression of mCHerry in BL21(DE3) most successful addition methods'' | ''Figure a3: pBAD expression of mCHerry in BL21(DE3) most successful addition methods'' | ||
| − | = GFP = | + | == pBAD expression of fluorophore GFP == |
| + | |||
| + | The results of all our tries are depicted in Figure b1 & b2 with the best results in b3. | ||
| + | |||
| + | [[File:T-TU-Eindhoven--pBAD_vector_GFP_60min.png]] | ||
| + | |||
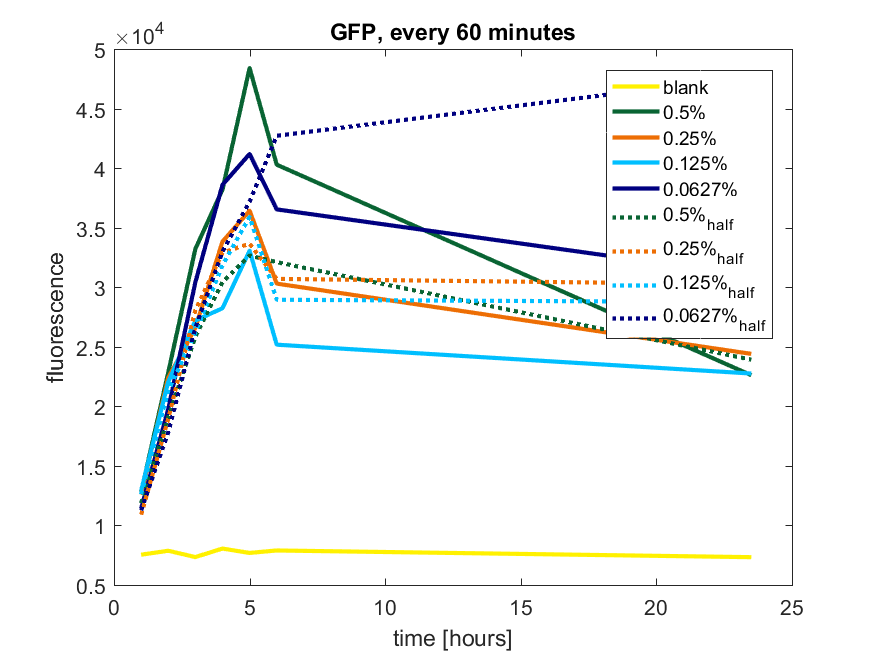
| + | ''Figure b1: pBAD expression of GFP in BL21(DE3) with every 60 minutes an addition of arabinose'' | ||
| + | |||
| + | [[File:T-TU-Eindhoven--pBAD_vector_GFP_90min.png]] | ||
| + | |||
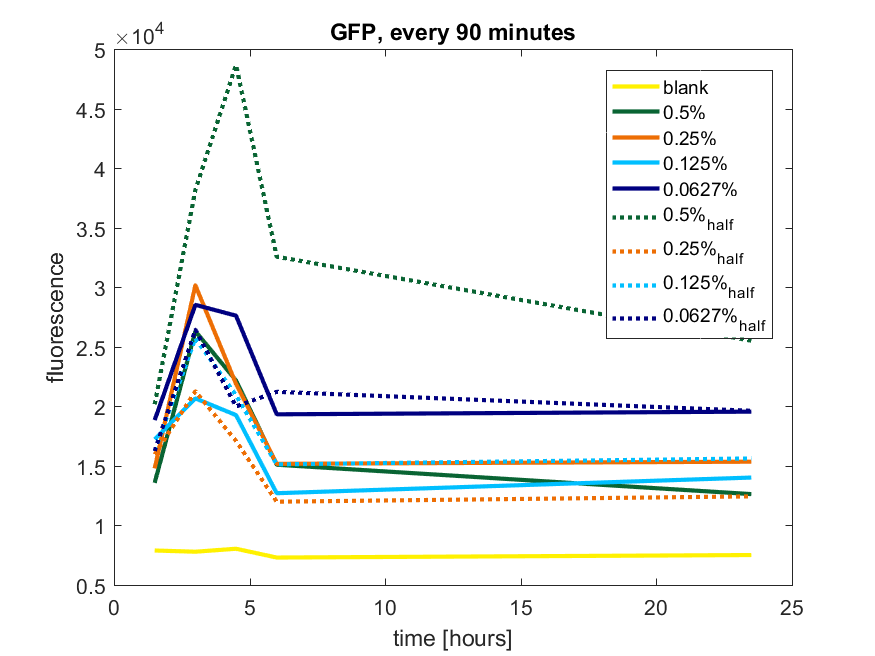
| + | ''Figure b2: pBAD expression of GFP in BL21(DE3) with every 90 minutes an addition of arabinose'' | ||
| − | |||
[[File:T-TU-Eindhoven--pBAD_vector_GFP_best_results.png]] | [[File:T-TU-Eindhoven--pBAD_vector_GFP_best_results.png]] | ||
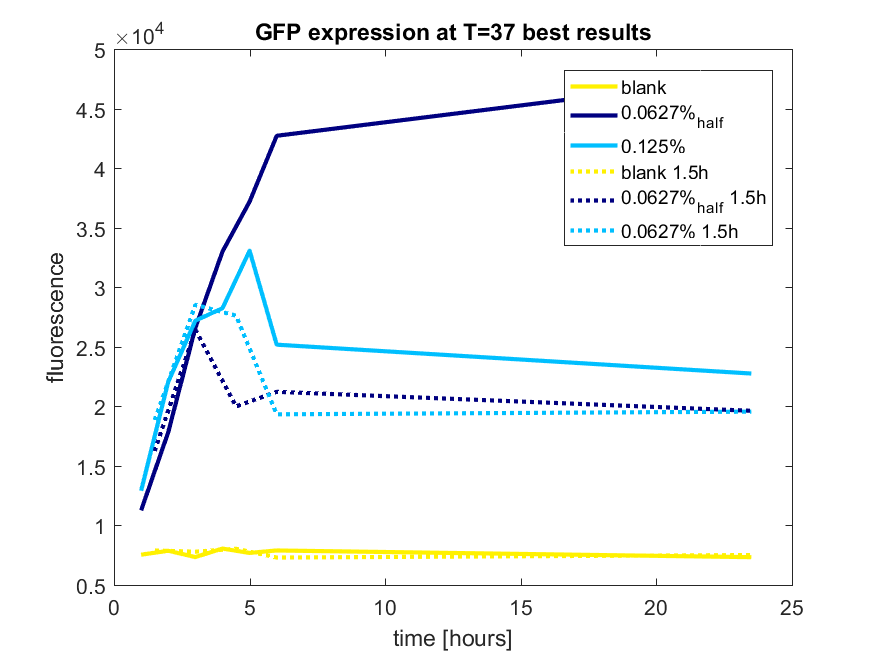
| − | ''Figure | + | ''Figure b3: pBAD expression of GFP in BL21(DE3) most successful addition methods'' |
| + | |||
| + | == pBAD expression conclusion == | ||
| + | |||
| + | The best results are depicted in Figure c. | ||
| + | |||
| + | [[File:T-TU-Eindhoven--pBAD_vector_mCherry_and_GFP_best_results.png]] | ||
| + | |||
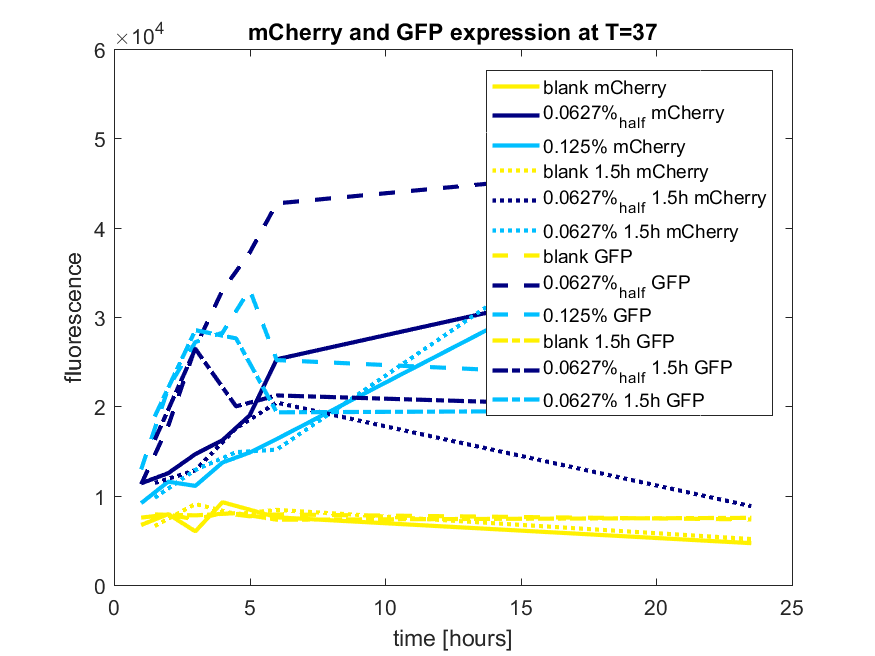
| + | ''Figure c: pBAD expression of mCherry and GFP in BL21(DE3) most successful addition methods'' | ||
= Advice = | = Advice = | ||
Revision as of 12:53, 28 October 2017
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_I0500
- When grown with 0.2% arabinose, promoter is weak-medium. [jb, 5/24/04] Part may not be compatible with MC4100 as cell line is araD 139.
- MC4100 is not a good chassis for operating BBa_I0500 (pBad promoter). The feed-forward regulation of the endogenous promoter controlling expression of the arabinose transporter prevents linear induction with increasing arabinose concentration. (Engineered strain from Keasling's lab, used by jrk for operation of the screening plasmid.) [cconboy 04]
- Observations of induced expression of GFP (BBa_E0840) are consistent with previous comments about weak promoter signal induction by jb 5/24/04. [melissali, Berkeley iGEM 2005]
PC and AraC are located on the complementary strand, reading right to left as written.
- At least one registry stock contains a deletion of the C at base 1194. This is after the transcriptional start but before the translation start, so it may not be significant. Parts with this mutation have been qualitatively observed to function normally.
- Can with success be combined with a promoter from pBAD SPL to obtain a very low leakiness and an appropriate strength. This is very efficient for expressing lethal proteins.
- Can be used for diauxic shifts with appropriate media as the promoter has a CAP binding site and thus shows catabolite repression with glucose [IISc Bangalore, 2016].
User Reviews
UNIQ05210c5063e5657d-partinfo-00000000-QINU
|
•••••
Antiquity |
This review comes from the old result system and indicates that this part worked in some test. |
|
iGEM Groningen 2009 |
The sequence was listed as inconsistent, and the ligations of parts behind the promoter failed. Restriction of isolated plasmids showed fragments of unexpected sizes. |
|
•••••
[http://2010.igem.org/Team:Slovenia 2010 iGEM team Slovenia] |
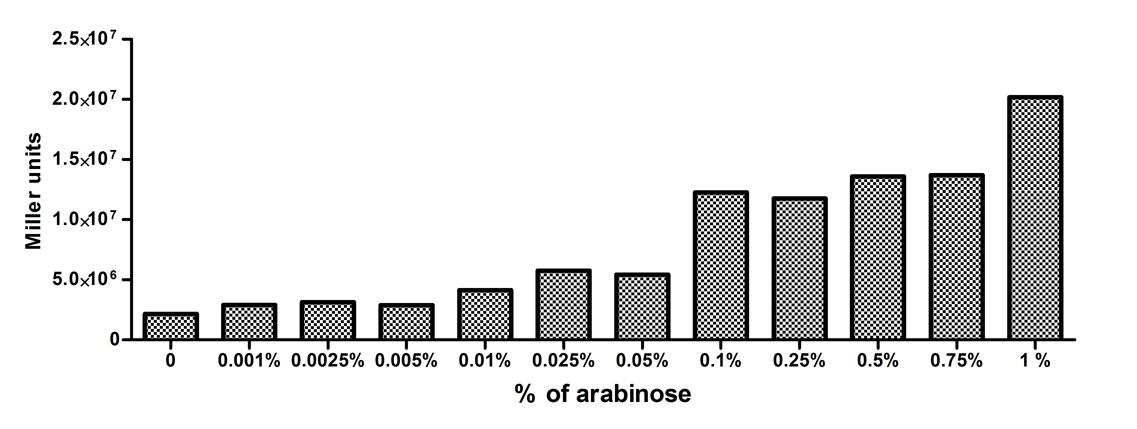
[http://2010.igem.org/Team:Slovenia 2010 iGEM team Slovenia] further characterized the pBAD/araC promoter (Part:Bba_I0500) using lacZ gene that encodes for beta-galactosidaze enzyme (Part:Bba_K323133). pBAD promoter is an arabinose inducible promoter. When incubating E.coli cultures containing the lacZ under pBAD/araC promoter, increasing concentrations of arabinose leads to higer promoter activity that results in higher amounts and subsequently higer activity of beta-galactosidase enzyme as seen from figure below. The resulting graph shows a trend of increasing beta-galactosidase activity over increasing concentrations of arabinose. The tested cultures incubated at different concentrations of arabinose (i.e. 0%, 0.001%, 0.0025%, 0.005%, 0.01%, 0.025%, 0.05%, 0.1%, 0.25%, 0.5%, 0.75% and 1%) have shown a trend of increasing beta-galactosidase activity with raising arabinose concentration. The activity was highest at 1% added arabinose, which implies the promotor should be induced by this arabinose concentration for reaching maximum expression of the regulated protein. However, some beta-galactosidase activity was observed even at 0% added arabinose, which indicates slight leakage of the pBAD promoter. pBAD/araC_lacZ_DTER construct (Part:Bba_K323133) includes a beta-galactosidase (lacZ) gene, the expression of which is regulated by the pBAD promoter (Part:Bba_I0500). This construct was designed for the sole purpose of characterising the pBAD promoter with the beta-galactosidase assay (for a detailed description of the method see the 2010 iGEM team Slovenia wiki). |
|
•••••
[http://2011.igem.org/Team:Groningen 2011 iGEM team Groningen] |
[http://2011.igem.org/Team:Groningen 2011 iGEM team Groningen] characterized the pBAD/AraC promoter (Part:Bba_I0500) even further using GFP and measuring the fluorescence in a fluorimeter over time, utilizing different arabinose concentrations to induce the promoter. After analysis of the obtained data (see graphs below), it is clear that the induction is faster and stronger with higher arabinose concentrations. It shows a dynamic response over a range of arabinose concentrations and over time. Because of that it can be treated as a precise tool capable of giving different output levels. The measurements were done in DH5alpha strain carrying a pSB1C3 with BBa_K607036 in triplicate.
|
|
•••••
[http://2011.igem.org/Team:Cambridge Cambridge 2011] |
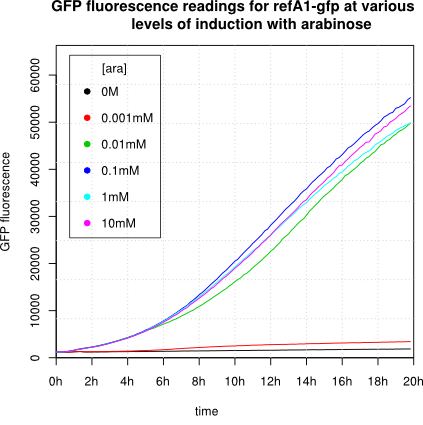
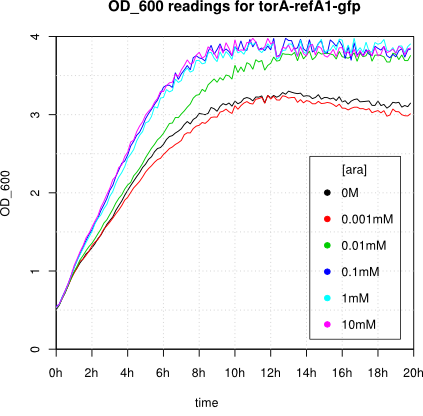
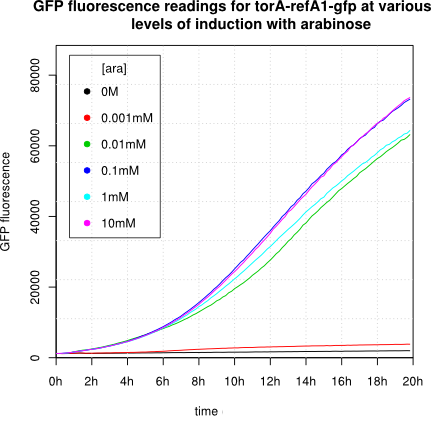
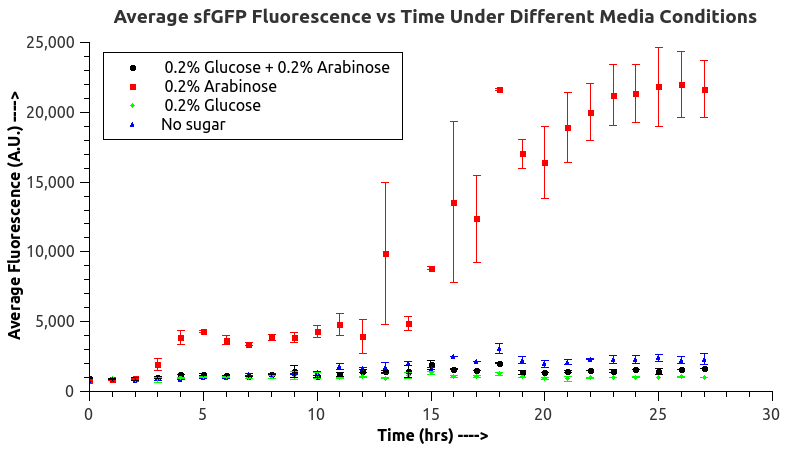
We observed an 'all-or-nothing' response in our use of the pBAD promoter. We grew up cells of the strain [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783] overnight at 37°C. We then diluted the culture 10-fold and grew the cells back up to an OD_600 of 0.5 to catch the cells in mid-exponential phase. We then induced with arabinose at various concentrations from 0.001mM up to 10mM and took OD and GFP readings on a plate reader over the next 20 hours. During this time the cells were held at 37°rees;C. The cells contained our ReflectinA1-sfGFP and TorA-ReflectinA1-sfGFP constructs on pSB3K3 plasmids. This work was not intended to characterise the pBAD promoter, but it highlights some of the difficulties in doing so:
Note: 1mM arabinose corresponds to 0.015% w/v.
|
|
[http://2012.igem.org/Team:UNITN-Trento Trento 2012] |
We were unable to extract this part from the distribution kit, so we developed our own! Please head to BBa_K731201 for documentation and characterization. |
|
••••
[http://2014.igem.org/Team: Hong_Kong_HKUST 2014] |
2014 HKUST iGEM team previously submitted [http://2014.igem.org/Team:Hong_Kong_HKUST/riboregulator/characterization characterization data] of Part BBa_I0500 that described an all-or-none behavior of the promoter. That is however not the case. We sincerely apologize for the misleading information. Please refer to our latest characterization data by HKUST-Rice 2015 below instead. UNIQ05210c5063e5657d-partinfo-0000000A-QINU |
|
•••••
[http://2015.igem.org/Team: HKUST-Rice 2015] |
[http://2015.igem.org/Team:HKUST-Rice HKUST-Rice Team] characterized BBa_I0500 using the construct BBa_I13540 in low copy plasmid pSB3K3. We aimed to obtain a dose response curve capturing the dynamic range and maximum induction concentration in order to compare its behavior when it is [http://2015.igem.org/Team:HKUST-Rice/Expression expressed in parallel with another sensor]. (please see BBa_K1682017 for more details. 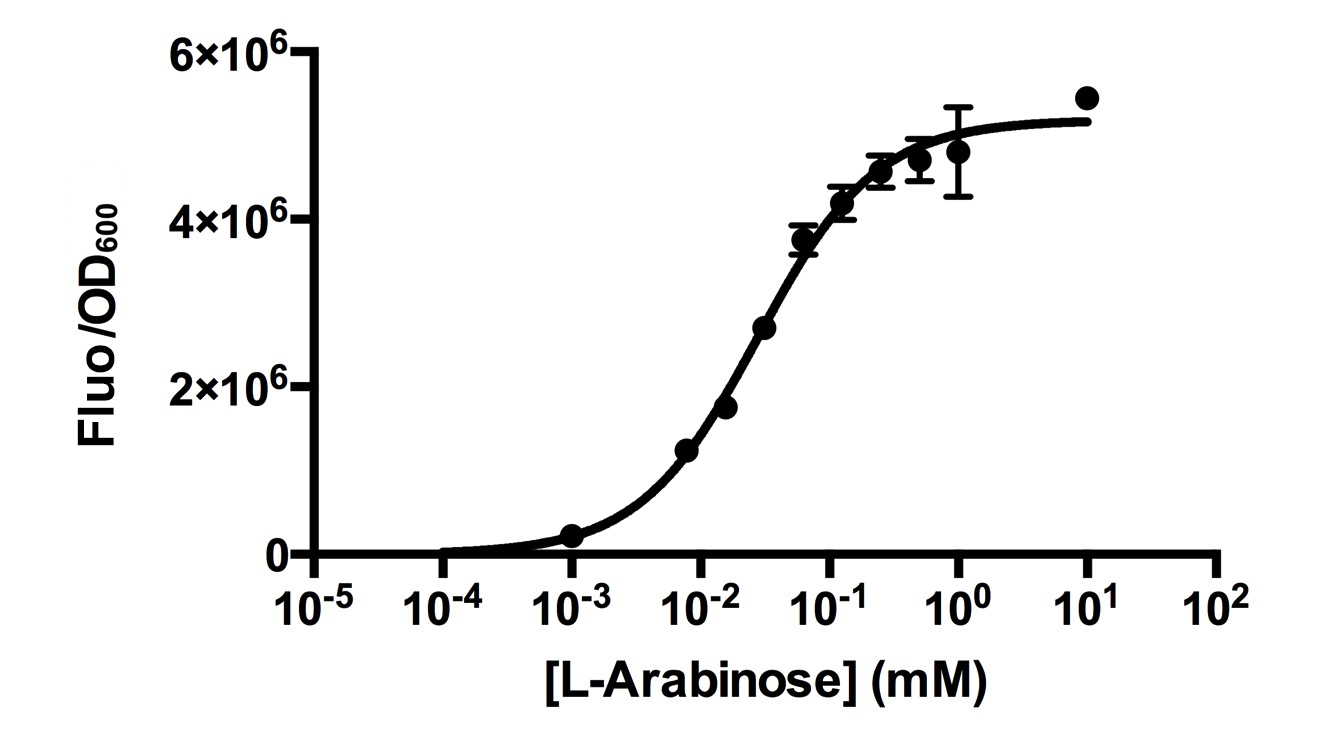 Figure 1. Dose response curve of cells carrying pSB3K3-BBa_I13540 under increasing L-arabinose concentration. Fluo = Relative Fluorescence Units. Error bars represent SD (n=3). Results indicate that the sensing range of BBa_I0500 lies at roughly 10-3 to 10-1 mM and has a half-maximal induction concentration of around 3-2mM of Arabinose.
Our results demonstrated that this promoter has a graded induction response and different sensing ranges on plasmids with different copy numbers (Figure 2). In concordance with literature, it displays an all-or-none behavior on a single cell level when expressed from a high copy plasmid pSB1K3, but not when expressed from a low copy plasmid pSB3K3 (Figure 3). 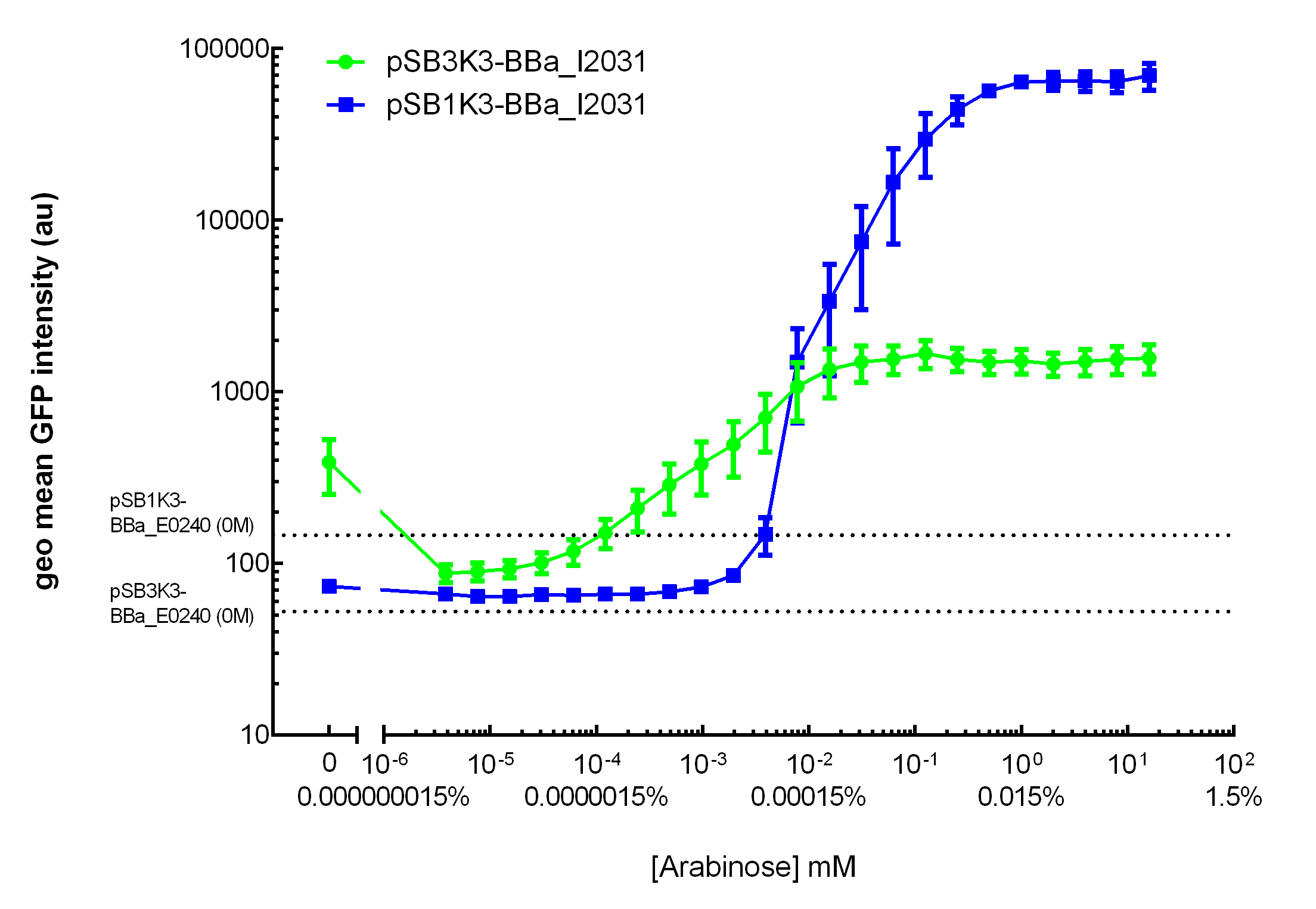 Figure 2. Transfer curves for BBa_I2031 on plasmid pSB3K3 and pSB1K3. On pSB3K3, BBa_I0500 is responsive to 10-4 - 10-2 mM arabinose, whereas on pSB1K3, it senses arabinose from roughly 10-3 to 1mM. These data agreed with neither Cambridge 2011 nor Groningen 2011’s result. The dashed lines represent cells’ auto fluorescence, measured using DH10B cells harboring pSB3K3-BBa_E0240 or pSB1K3-BBa_E0240. Error bars represent SEM of 3 independent experiments on 3 different days. The molar concentration to percentage conversion was provided for comparison with previous data. 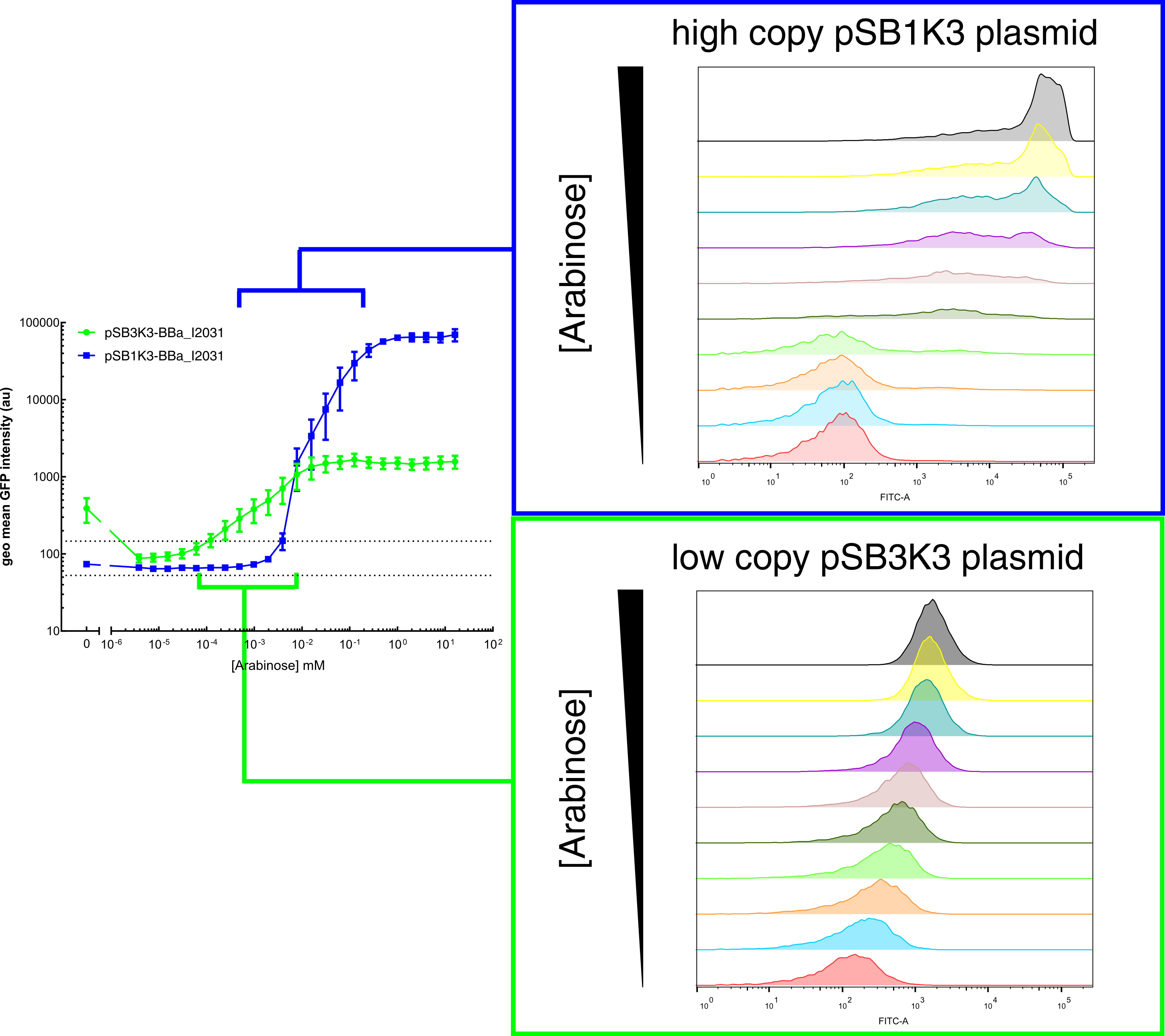 Figure 3. Histogram plots for sensing ranges of BBa_I0500 on high and low copy plasmid. An all-or-none induction can be observed when BBa_I0500 is placed on a high copy plasmid, where induced cells were mostly distributed among two bins of fluorescence. Yet on low copy plasmid, the induced populations remain homogenous along the arabinose concentration gradient. Concentrations of arabinose for high copy pSB1K3 plasmid: 0.488µM – 0.25mM. For low copy pSB3K3 plasmid: 0.0610µM – 0.03125mM. Only 1 set of experiment result from 3 replicates is presented. Moreover, we would like to warn potential users of this part not to drive expression of riboregulators ([http://www.ncbi.nlm.nih.gov/pubmed/15208640 Isaacs et al., 2004]) using BBa_I0500. This PBAD promoter contains a 19bp sequence after the transcription start site, which would produce a taRNA with additional nucleotides at its 5’ end, and rendering it incapable to activate crRNA. To avoid misuse of this part due to incognizance of its sequence information, we recommend users to look up BBa_K1067007 for annotations. For more information and methods of our characterization, please refer to our detailed characterization report available through our [http://2015.igem.org/Team:HKUST-Rice/Expression/ParaBAD wiki page]. |
|
•••••
[http://2016.igem.org/Team:IISc_Bangalore]Team IISc Bangalore |
IISc Bangalore characterized BBa_I0500 by using Bba_746908 which has sfGFP under the pBAD/araC promoter. We wanted to verify the catabolite repression of the expression from the promoter in the presence of glucose. |
|
•••••
[http://2016.igem.org/Team:OUC-China]Team OUC-China |
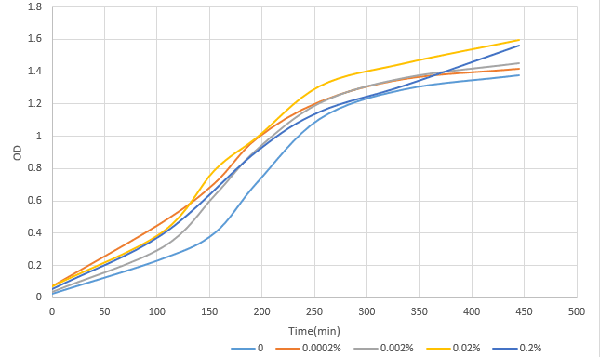
[http://2016.igem.org/Team:OUC-China OUC-China] characterized BBa_I0500 of different concentrations of L-arabinose on the transcriptional level. We found that the strength of I0500 was mainly measured on the translational level before. As far as we are concerned, it is the transcriptional level that can directly reflect I0500’s original strength without interfering by context. In order to verify it more accurately, we tested it on the transcriptional level by extracting RNA and measured it by qPCR. We constructed I0500 with GFP(E0040) and transformed into TOP10F’. Firstly , we plated on LB agar with chloramphenicol and individual colonies were subsequently grown overnight in LB-medium. Cultures were back diluted 1:100 into 100mL fresh LB-medium with chloramphenicol, adding 0,0.0002%,0.002%,0.02%,0.2% (m/V)L-arabinose. After growing for 18 hours, we extracted RNA and measured it. At the same time, we measured the fluorescence of GFP(485nm/520nm). Besides, we also measured OD over time to check whether L-arabinose promotes bacteria’s growth. 1. 100mlLB,blank |
Contribution iGEM TU-Eindhoven 2017 http://2017.igem.org/Team:TU-Eindhoven]
Method
We first tested three different temperatures, being 20, 25 and 37 degree Celsius. From our fist basic measurements, we discovered that expression was more efficient at a temperature of 37 degree Celsius and continued with this setting on our incubator. The next step was consequent adding arabinose to the cultures, as E. coli BL21(DE3) is a strain that can break down arabinose, and for an optimal expression, it is better to have a high enough constant concentration. We tested an addition of every 60 minutes and every 90 minutes, with varying concentrations of arabinose. The percentages that are depicted in the figures are the total concentration of arabinose in the whole culture.
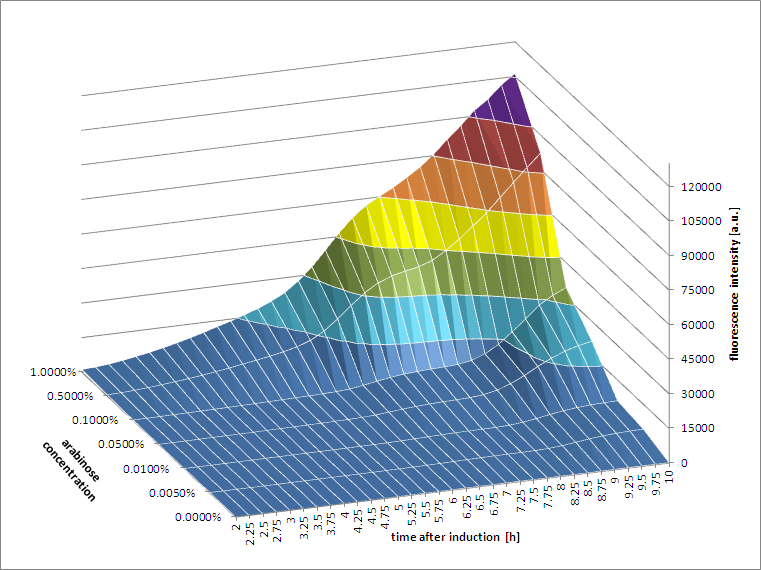
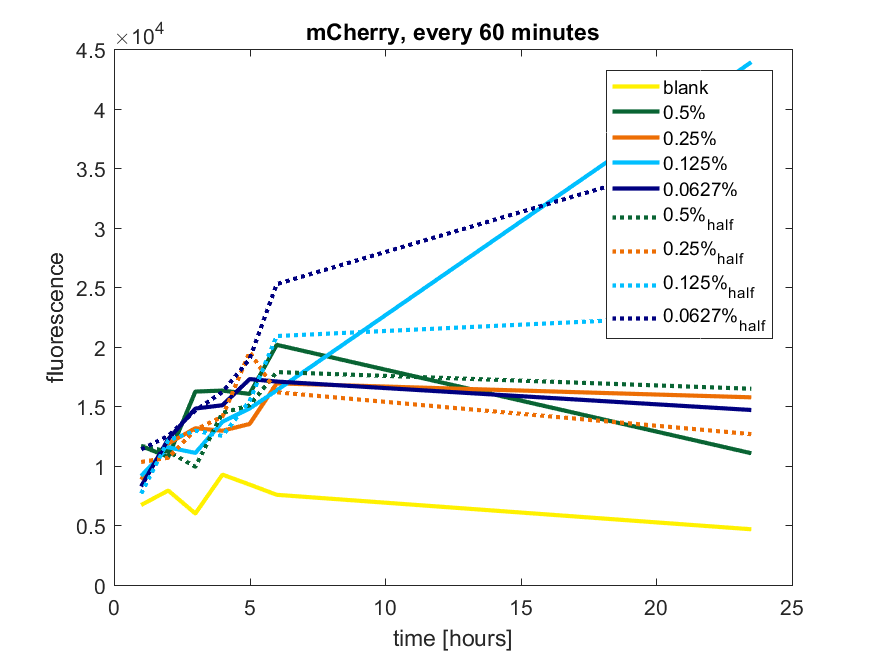
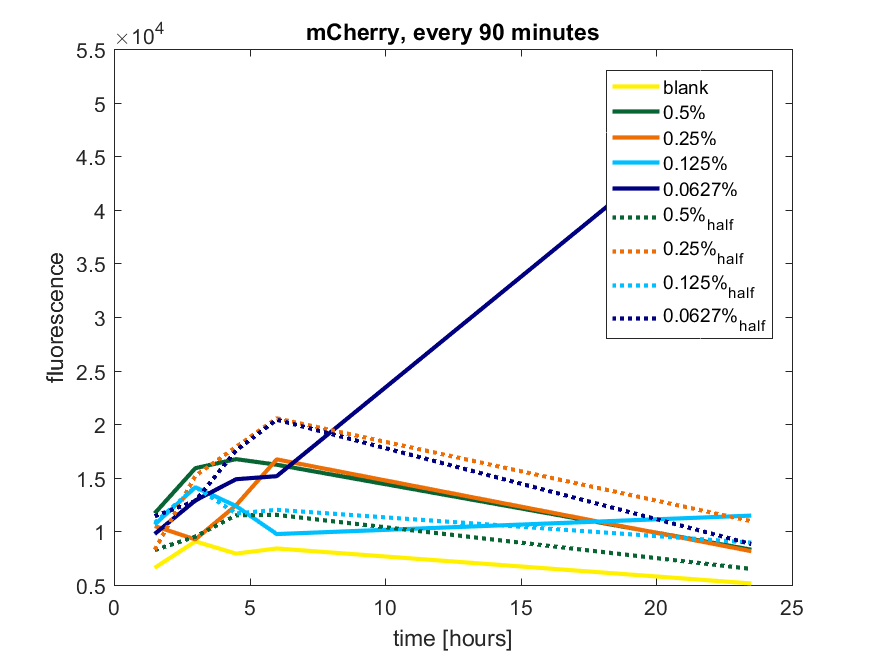
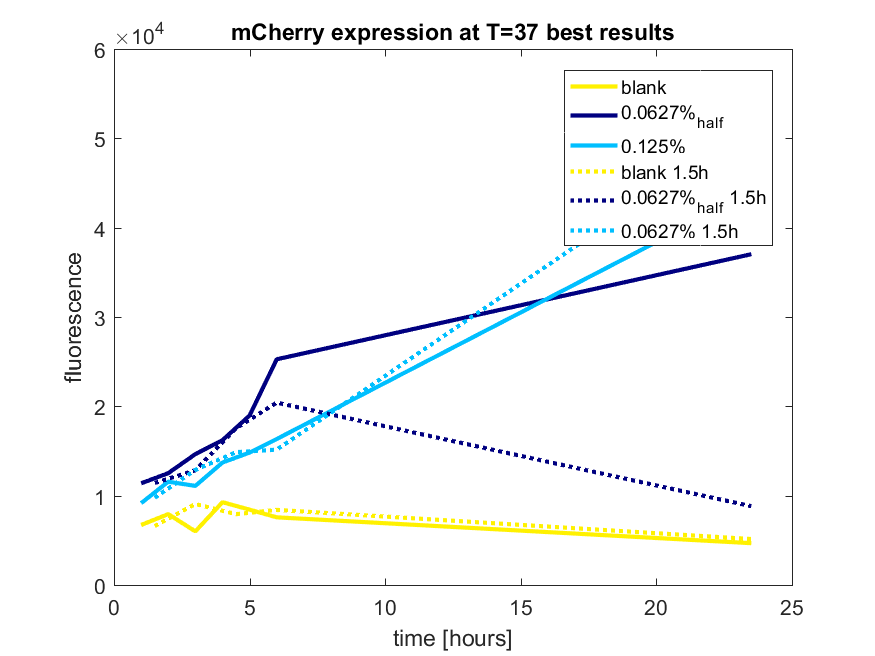
pBAD expression of fluorophore mCherry
The results of all our tries are depicted in Figure a1 & a2 with the best results in a3.
Figure a1: pBAD expression of mCherry in BL21(DE3) with every 60 minutes an addition of arabinose
Figure a2: pBAD expression of mCherry in BL21(DE3) with every 90 minutes an addition of arabinose
Figure a3: pBAD expression of mCHerry in BL21(DE3) most successful addition methods
pBAD expression of fluorophore GFP
The results of all our tries are depicted in Figure b1 & b2 with the best results in b3.
Figure b1: pBAD expression of GFP in BL21(DE3) with every 60 minutes an addition of arabinose
Figure b2: pBAD expression of GFP in BL21(DE3) with every 90 minutes an addition of arabinose
Figure b3: pBAD expression of GFP in BL21(DE3) most successful addition methods
pBAD expression conclusion
The best results are depicted in Figure c.
Figure c: pBAD expression of mCherry and GFP in BL21(DE3) most successful addition methods
Advice
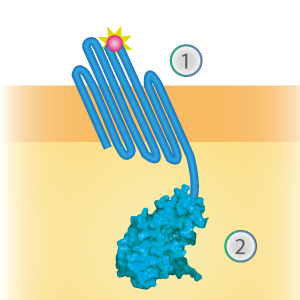
LargeBit is the big part of this Split Luciferase. Its molecular weight is 18 kDa. OmpX (1) (with a correct mutation for the amber stop codon TAG), a BamHI-linker and LargeBit (2) together will look like Figure 1. This construct on its own has no function since LargeBit on itself has no luminescence activity.
Figure 1: Schematical overview of the OmpX - LargeBit construct.