Difference between revisions of "Part:BBa K2217001"
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K2217001 short</partinfo> | <partinfo>BBa_K2217001 short</partinfo> | ||
| + | |||
RNA sequence was retrieved from circ-Interactome database Regulatory circular RNA hsa-circ-0000064 used in Competing endogenous RNA ceRNA circuit, We used this part as a competing endogenuous RNA to regulate Hepatocellular-Carcinoma associated miRNA mir-1825. | RNA sequence was retrieved from circ-Interactome database Regulatory circular RNA hsa-circ-0000064 used in Competing endogenous RNA ceRNA circuit, We used this part as a competing endogenuous RNA to regulate Hepatocellular-Carcinoma associated miRNA mir-1825. | ||
| Line 17: | Line 18: | ||
| − | |||
| − | |||
| + | ===Usage and Biology=== | ||
| + | RNA (hsa-circ-0000064) was submitted to the registry after biomarker filtration using bioinformatics databases demonstrated at our biomarker filtration at [http://2017.igem.org/Design.html# Egypt-AFCM Design], to be used as a competing endogenous RNA in a ceRNA network. | ||
<!-- --> | <!-- --> | ||
Revision as of 17:55, 22 October 2017
Circular RNA hsa-circ-000064
RNA sequence was retrieved from circ-Interactome database Regulatory circular RNA hsa-circ-0000064 used in Competing endogenous RNA ceRNA circuit, We used this part as a competing endogenuous RNA to regulate Hepatocellular-Carcinoma associated miRNA mir-1825.
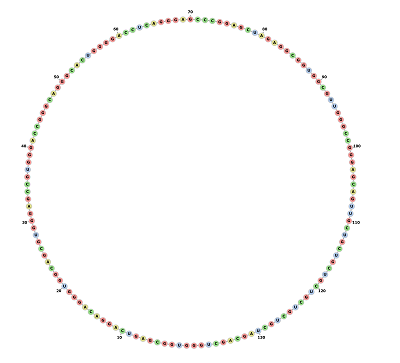
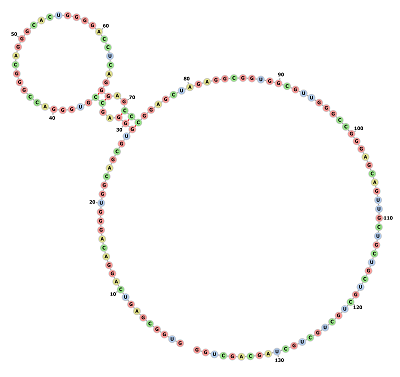
Circular RNA Model
Minimum free energy prediction using RNAFOLD generated an optimal secondary structure in dot-bracket notation from a centroid structure of 0.00 kcal/mol minimum free energy Structure
to 1.78 kcal/mol minimum free energy Structure
More Information about circular design of this part can be found at [http://2017.igem.org/Modeling.html# Egypt-AFCM Modeling]
Usage and Biology
RNA (hsa-circ-0000064) was submitted to the registry after biomarker filtration using bioinformatics databases demonstrated at our biomarker filtration at [http://2017.igem.org/Design.html# Egypt-AFCM Design], to be used as a competing endogenous RNA in a ceRNA network.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 128
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]