Difference between revisions of "Part:BBa K1893013"
| Line 9: | Line 9: | ||
STAR is is the first instance of small bacterial RNAs being used to directly activate transcription. STAR binds a sense target RNA sequence, the pAD1 plasmid attenuator sequence, which is fused upstream of the gene of interest. The pAD1 plasmid attenuator sequence forms an intrinsic hairpin structure, preventing RNA polymerase from transcribing the gene and thus acting as a terminator of transcription. However, when STAR is produced, it binds to the target sequence and interferes with the hairpin formation, allowing transcription of the downstream gene. | STAR is is the first instance of small bacterial RNAs being used to directly activate transcription. STAR binds a sense target RNA sequence, the pAD1 plasmid attenuator sequence, which is fused upstream of the gene of interest. The pAD1 plasmid attenuator sequence forms an intrinsic hairpin structure, preventing RNA polymerase from transcribing the gene and thus acting as a terminator of transcription. However, when STAR is produced, it binds to the target sequence and interferes with the hairpin formation, allowing transcription of the downstream gene. | ||
| − | Using STAR offers significant advantages over traditional protein-based regulatory systems, including: | + | <b>Using STAR offers significant advantages over traditional protein-based regulatory systems, including:<b> |
| − | + | ||
→ Low metabolic burden: the RNA is functional after transcription, avoiding the resource-intensive translation steps. <br> | → Low metabolic burden: the RNA is functional after transcription, avoiding the resource-intensive translation steps. <br> | ||
→ Portability: trans-acting RNA based regulation is found throughout the bacterial kingdom (as well as other kingdoms) and is not host-specific. <br> | → Portability: trans-acting RNA based regulation is found throughout the bacterial kingdom (as well as other kingdoms) and is not host-specific. <br> | ||
Revision as of 15:32, 27 October 2016
Small transcription activating RNA (STAR)
This is the STAR (Small Transcription Activating RNA) construct with its own transcription terminator t500. It is a novel RNA-logic technology that allows for rapid and robust regulation of gene transcription. When transcribed, it produces the STAR-antisense RNA (pAD1.A5), which can bind to the sense STAR-target terminator attenuator sequence (pAD1.S5). Thus, only in the presence of STAR-antisense, transcription and expression of coding sequences downstream to the STAR-target is activated.
Usage and Biology
STAR is is the first instance of small bacterial RNAs being used to directly activate transcription. STAR binds a sense target RNA sequence, the pAD1 plasmid attenuator sequence, which is fused upstream of the gene of interest. The pAD1 plasmid attenuator sequence forms an intrinsic hairpin structure, preventing RNA polymerase from transcribing the gene and thus acting as a terminator of transcription. However, when STAR is produced, it binds to the target sequence and interferes with the hairpin formation, allowing transcription of the downstream gene.
Using STAR offers significant advantages over traditional protein-based regulatory systems, including:<b>
→ Low metabolic burden: the RNA is functional after transcription, avoiding the resource-intensive translation steps.
→ Portability: trans-acting RNA based regulation is found throughout the bacterial kingdom (as well as other kingdoms) and is not host-specific.
→ Tight Regulation: In the absence of STAR there is very low baseline expression of the downstream gene, while in the presence of STAR the expression is activated 96-fold.
→ Ease of design: Watson-Crick base pairing avoids the need for structural protein-protein interactions, which are difficult to predict and design.
→ Reduced lag in circuits: RNA degradation is rapid, reducing the lag in the comparison function, and offering short time response for our comparator circuit.
Characterisation data
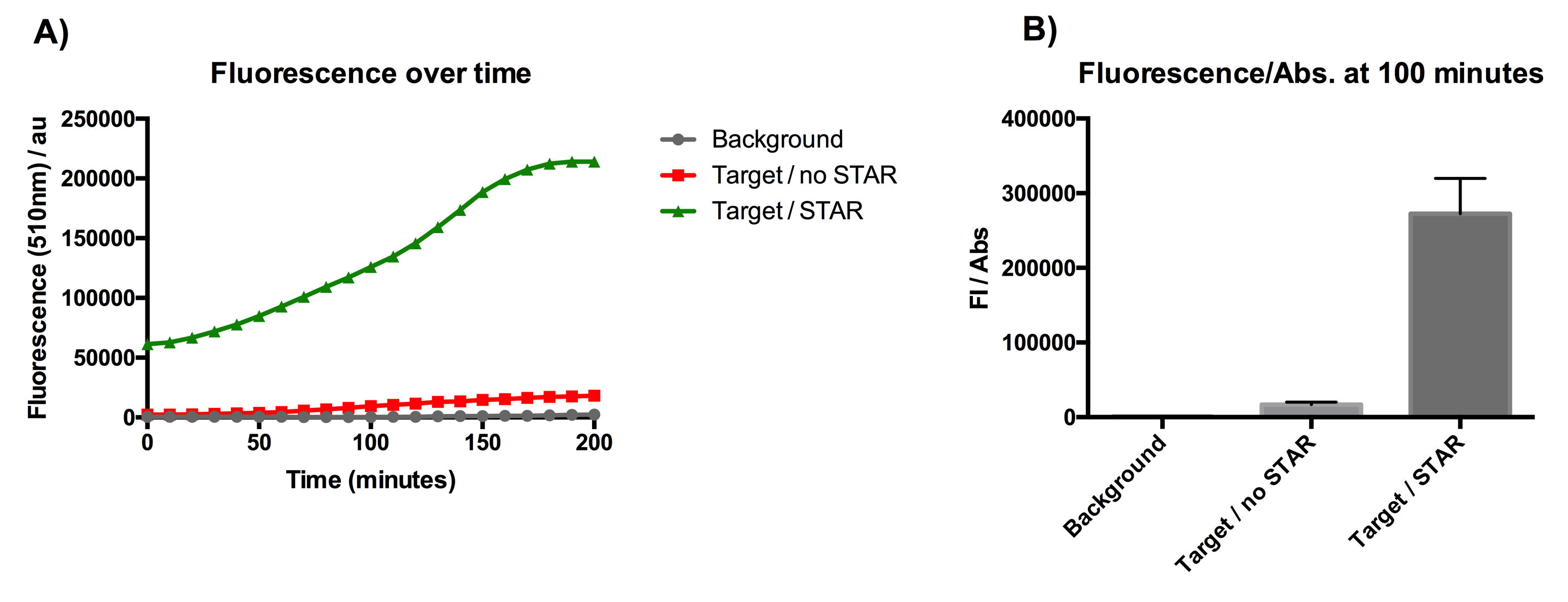
Figure 1: Characterisation of STAR system in TOP10 E. coli cells. (A) Normalised fluorescence monitored over time for cell lines incorporating the STAR system in the absence or presence of transcribed STAR molecules (B) Normalised endpoint fluorescence (100 minutes) for cell lines in the absence or presence of STAR molecules. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. Normalised fluorescence was calculated by dividing fluorescent signal by the O.D.600 value of the culture. Background was determined by the use of DH10B cells with no plasmid transformed. Error bars represent standard deviation from 3 technical repeats.
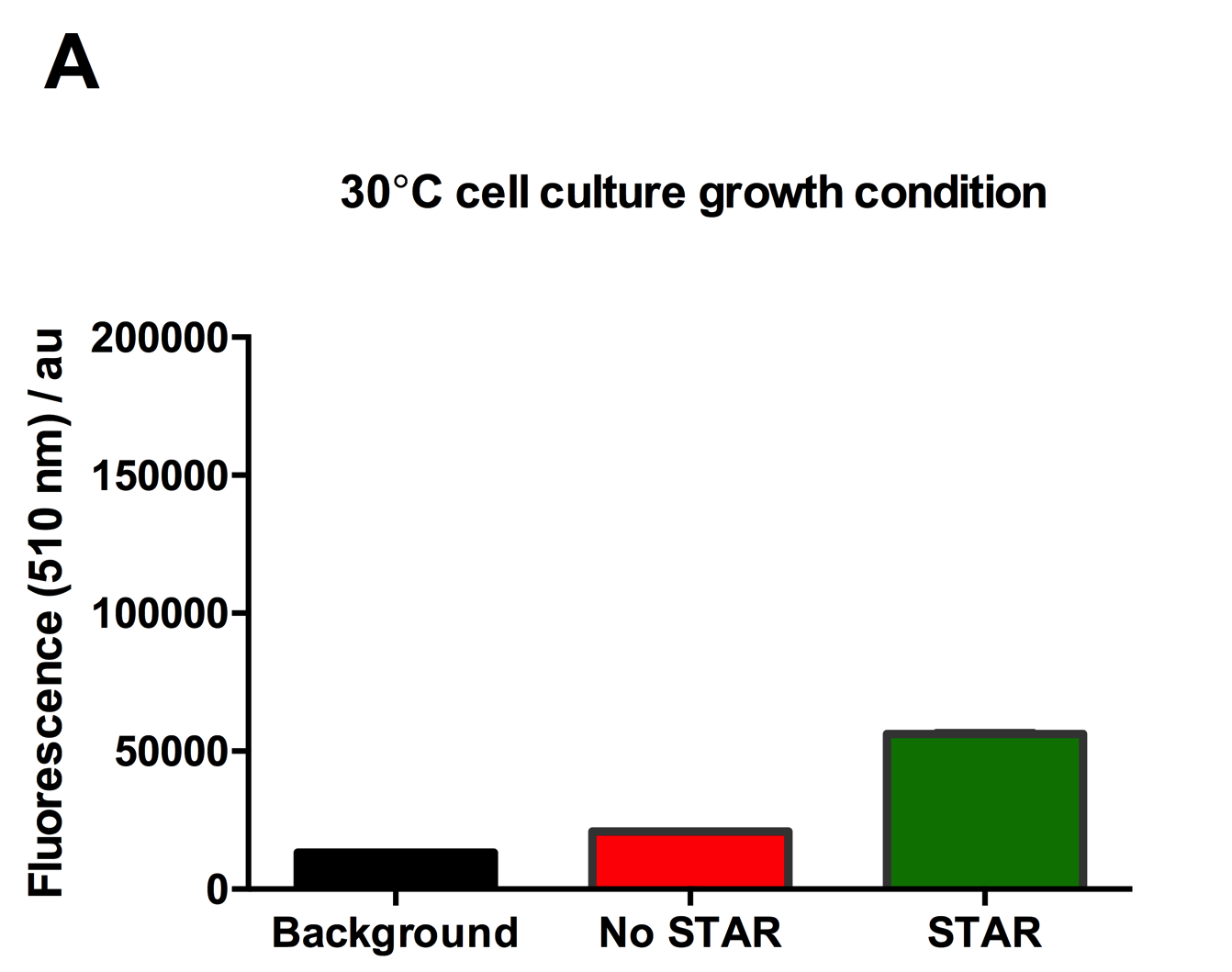
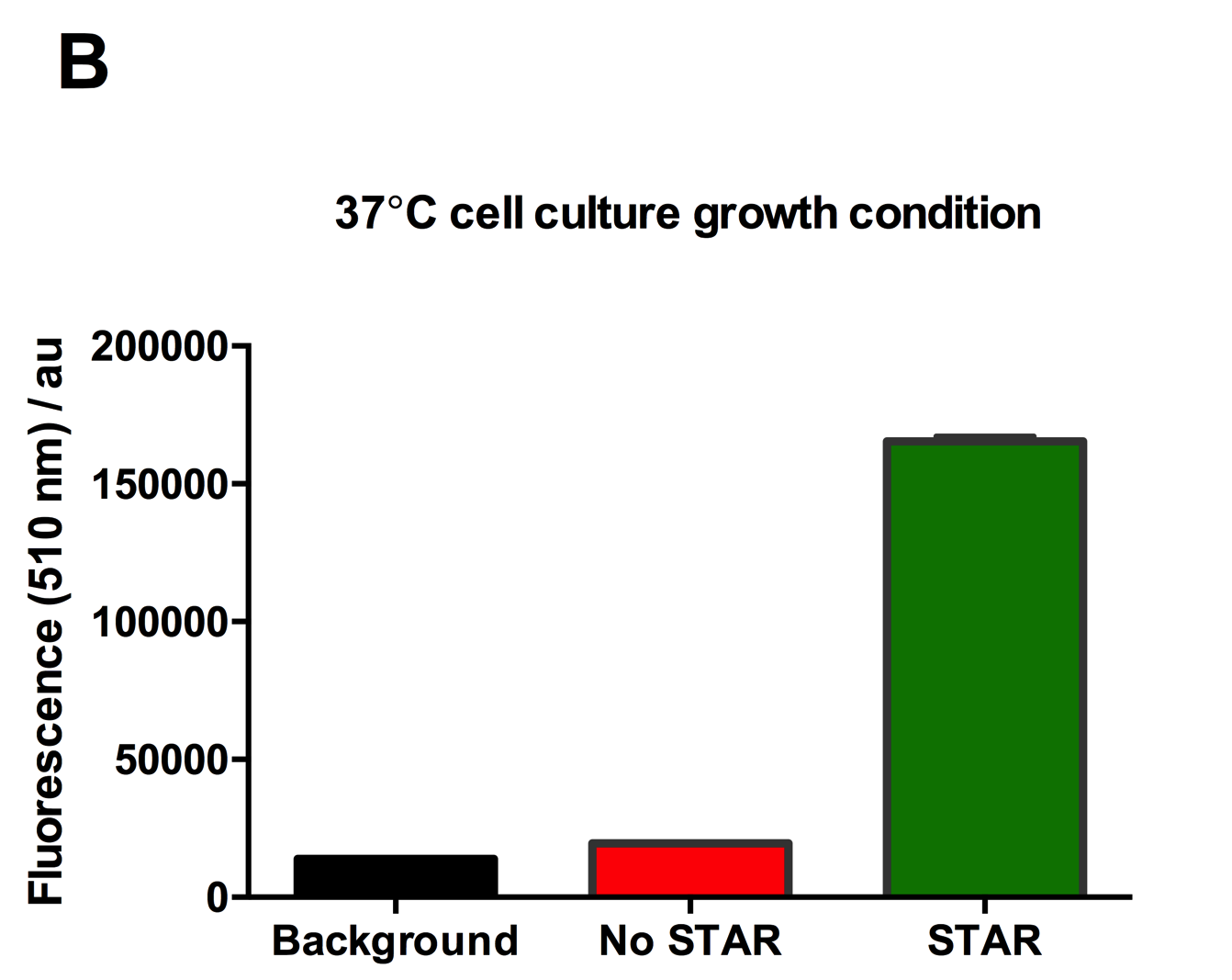
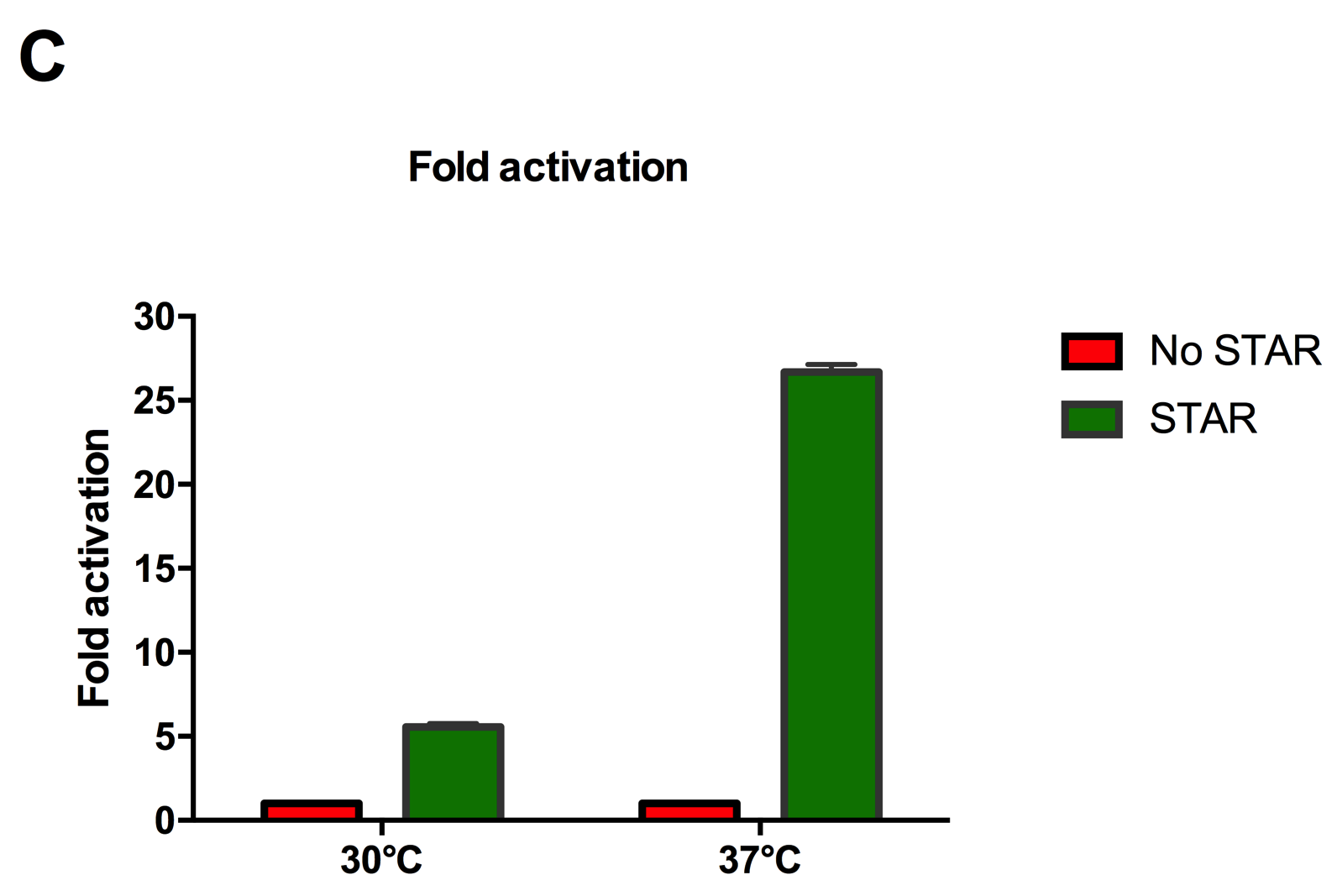
Figure 2: Characterisation of STAR system in TOP10 E. coli cells at different temperatures. (A) Cell culture fluorescence at 30°C (B) Cell culture fluorescence assay at 37°C. (C) Fold activation SFGFP expression in presence of STAR. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. The autofluorescence background control used is E. coli Top 10 cells with no reporter plasmid. Error bars represent standard deviation from 3 technical repeats.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]