Difference between revisions of "Part:BBa J23112"
(→Note) |
|||
| Line 22: | Line 22: | ||
==Note== | ==Note== | ||
Parts <partinfo>J23103</partinfo> and <partinfo>J23112</partinfo> are the same (Rahmi Lale). | Parts <partinfo>J23103</partinfo> and <partinfo>J23112</partinfo> are the same (Rahmi Lale). | ||
| + | |||
| + | ===iGEM Team 2019 Stuttgart Characterization=== | ||
| + | <html> | ||
| + | <br/> | ||
| + | We added quantitative experimental characterization to the following biobricks: <br> | ||
| + | <a href="https://parts.igem.org/Part:BBa_J23100">BBa_J23100</a> | ||
| + | <a href="https://parts.igem.org/Part:BBa_J23112">BBa_J23112</a> | ||
| + | <a href="https://parts.igem.org/Part:BBa_J23118">BBa_J23118</a> | ||
| + | <br/> | ||
| + | These parts are a family of constitutive promoters. They were present in plasmid <a | ||
| + | href="https://parts.igem.org/Part:BBa_J61002">BBa_J61002</a>. | ||
| + | We analyzed the influence of the mentioned promoters on the expression of red fluorescence protein (RFP), | ||
| + | both on protein level (fluorescence spectroscopy) and mRNA level using reverse transcriptase quantitative polymerase | ||
| + | chain reaction (RT-qPCR). | ||
| + | <p> | ||
| + | Biobricks were transformed in <i>E. coli</i> MG1655. Cells were harvested in the early exponential phase and | ||
| + | fluorescence was measured. Also, 500 µL of culture was centrifuged and the pellet was used for RNA extraction. | ||
| + | In addition to that, <i>E. coli</i> MG1655 was cultured and harvested for RNA isolation without the plasmid | ||
| + | BBa_J61002 to ensure, that our primers specifically bind to RFP mRNA. <br/> | ||
| + | <br/> | ||
| + | RT-qPCR was performed and analyzed using the ddCt method. To quantify the expression levels of RFP under different | ||
| + | promoters, the cDNA of a house-keeping gene was amplified as well. We chose a GTPase-activation protein (GAP) as a | ||
| + | housekeeping gene. <br/> | ||
| + | <br/> | ||
| + | <br> | ||
| + | <strong>Results of fluorescence spectroscopy</strong><br> | ||
| + | <br> | ||
| + | Investigating the grown cell cultures by fluorescence spectroscopy clearly showed emission at 590nm (excitation at | ||
| + | 540 nm) indicating a successful expression of RFP. Cells containing the promoter BBa_J23100 showed an emission at | ||
| + | 590 nm of 192 +- 13. Compared to that, cells containing the promoter BBa_J23112 showed an emission of 30 +- 1. The | ||
| + | highest emission was shown by cells containing the promoter BBa_J23118: 345 +- 4. | ||
| + | <br> | ||
| + | <br> | ||
| + | <strong>Results of qPCR</strong><br> | ||
| + | <br> | ||
| + | Figure 1 shows, that RFP expression was about 7-fold higher than the expression of GAP in cells containing promoter BBa_J23100. Compared to | ||
| + | that, the expression of RFP in cells containing promoter BBa_J23112 was about 55-fold higher than expression of the | ||
| + | house-keeping gene. The highest expression of RFP compared to GAP was observed in cells containing promoter BBa_J23118. | ||
| + | Here, RFP expression was 74-fold compared to GAP expression. | ||
| + | <br> | ||
| + | |||
| + | <img style="width:50%; display: block" | ||
| + | src="https://2019.igem.org/wiki/images/thumb/5/50/T--Stuttgart--balkendiagramm_charaktereisierung.png/800px-T--Stuttgart--balkendiagramm_charaktereisierung.png"/> | ||
| + | <small>Figure 1: Average relative quantification of RFP expression compared to the expression of the house-keeping gene GAP in <i>E. coli</i> | ||
| + | MG1655. | ||
| + | Cells contained the plasmid BBa_J61002 with different promoters: BBa_J23100, BBa_J23112 and BBa_J23118. As a | ||
| + | control | ||
| + | <i>E. coli</i> MG1655 were cultivated without the plasmid. RT-qPCR measurements were carried out in technical triplicates. Standard deviation is shown</small> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | For more information, detailed material and method description, please visit our <a href="https://2019.igem.org/Team:Stuttgart/Contribution">wiki</a> | ||
| + | </html> | ||
Revision as of 20:41, 20 October 2019
constitutive promoter family member
ATTENTION: This part is a duplicate of BBa_J23103. --mosthege 05:56, 3 April 2015 (CDT)
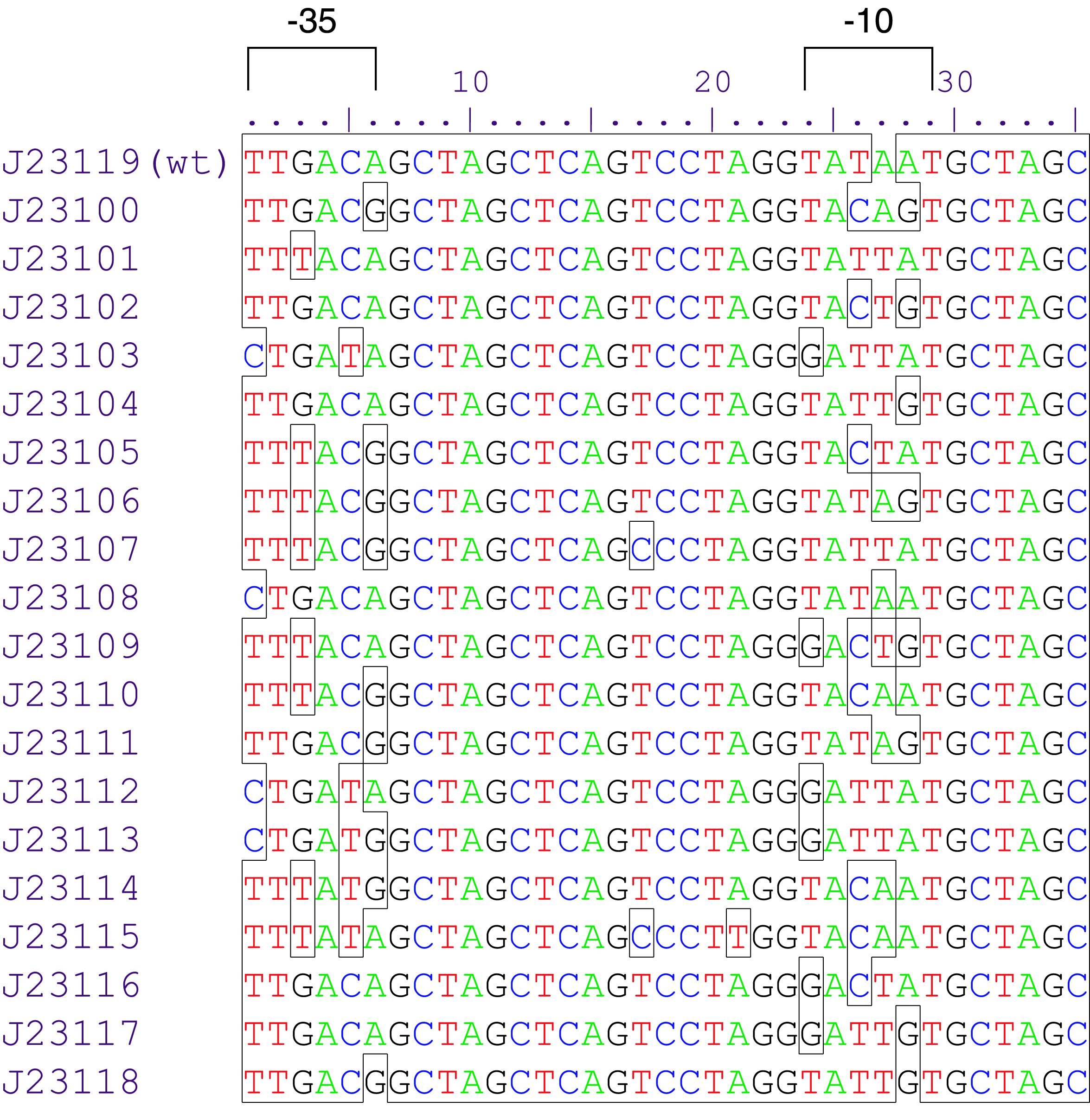
Variant RFP (au) J23112 1 J23103 17 J23113 21 J23109 106 J23117 162 J23114 256 J23115 387 J23116 396 J23105 623 J23110 844 J23107 908 J23106 1185 J23108 1303 J23118 1429 J23111 1487 J23101 1791 J23104 1831 J23102 2179 J23100 2547 |
Constitutive promoter family
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part BBa_J61002 for details on their use.
These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. JCAraw
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Note
Parts BBa_J23103 and BBa_J23112 are the same (Rahmi Lale).
iGEM Team 2019 Stuttgart Characterization
We added quantitative experimental characterization to the following biobricks:
BBa_J23100
BBa_J23112
BBa_J23118
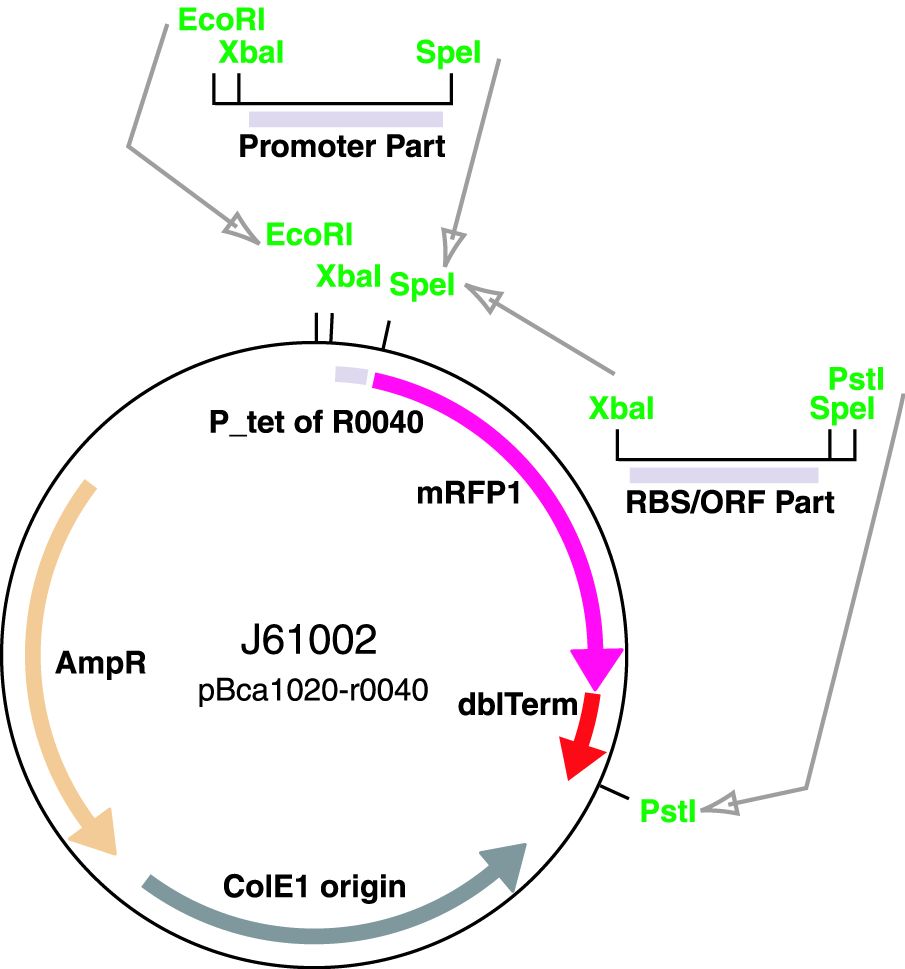
These parts are a family of constitutive promoters. They were present in plasmid BBa_J61002.
We analyzed the influence of the mentioned promoters on the expression of red fluorescence protein (RFP),
both on protein level (fluorescence spectroscopy) and mRNA level using reverse transcriptase quantitative polymerase
chain reaction (RT-qPCR).
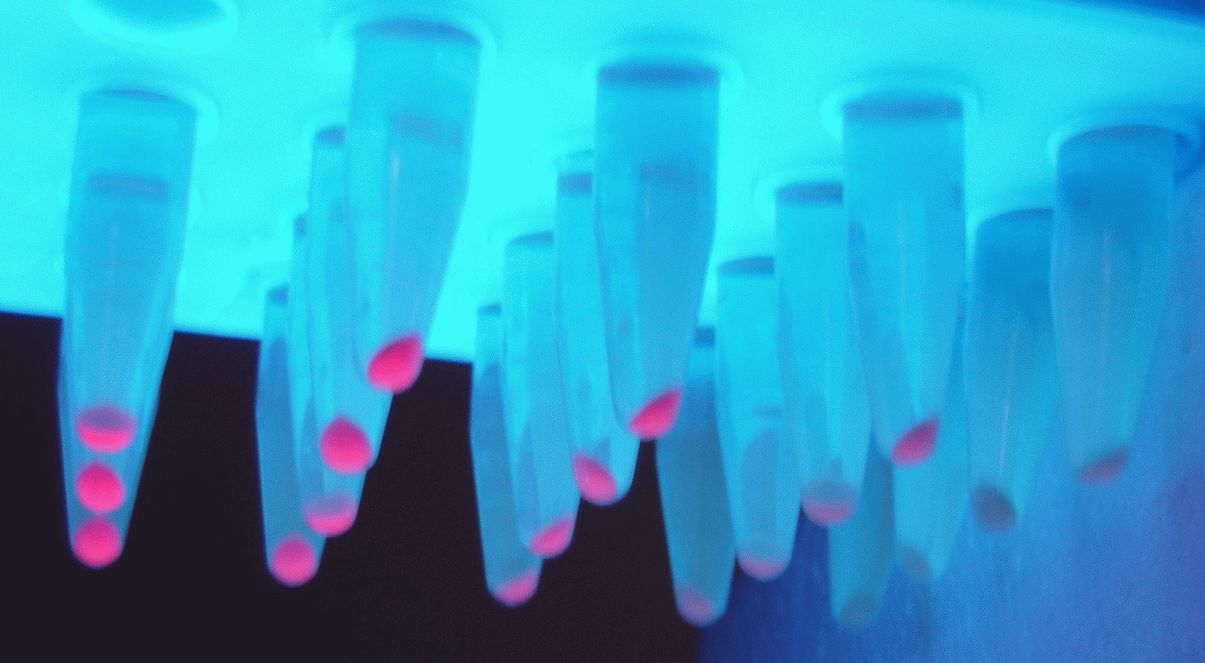
Biobricks were transformed in E. coli MG1655. Cells were harvested in the early exponential phase and
fluorescence was measured. Also, 500 µL of culture was centrifuged and the pellet was used for RNA extraction.
In addition to that, E. coli MG1655 was cultured and harvested for RNA isolation without the plasmid
BBa_J61002 to ensure, that our primers specifically bind to RFP mRNA.
RT-qPCR was performed and analyzed using the ddCt method. To quantify the expression levels of RFP under different
promoters, the cDNA of a house-keeping gene was amplified as well. We chose a GTPase-activation protein (GAP) as a
housekeeping gene.
Results of fluorescence spectroscopy
Investigating the grown cell cultures by fluorescence spectroscopy clearly showed emission at 590nm (excitation at
540 nm) indicating a successful expression of RFP. Cells containing the promoter BBa_J23100 showed an emission at
590 nm of 192 +- 13. Compared to that, cells containing the promoter BBa_J23112 showed an emission of 30 +- 1. The
highest emission was shown by cells containing the promoter BBa_J23118: 345 +- 4.
Results of qPCR
Figure 1 shows, that RFP expression was about 7-fold higher than the expression of GAP in cells containing promoter BBa_J23100. Compared to
that, the expression of RFP in cells containing promoter BBa_J23112 was about 55-fold higher than expression of the
house-keeping gene. The highest expression of RFP compared to GAP was observed in cells containing promoter BBa_J23118.
Here, RFP expression was 74-fold compared to GAP expression.
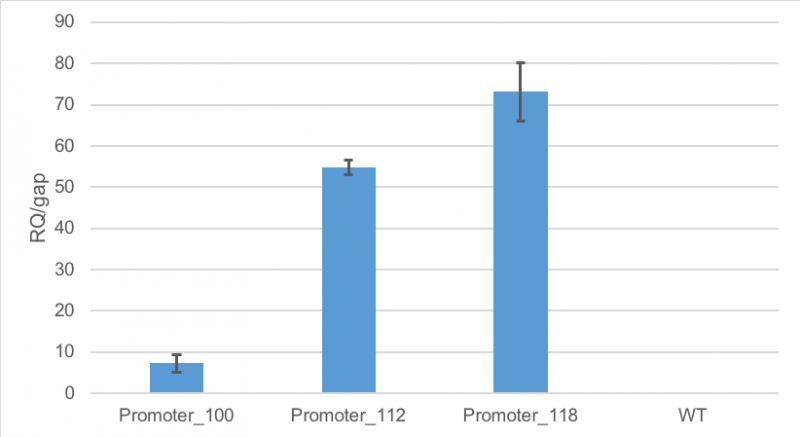 Figure 1: Average relative quantification of RFP expression compared to the expression of the house-keeping gene GAP in E. coli
MG1655.
Cells contained the plasmid BBa_J61002 with different promoters: BBa_J23100, BBa_J23112 and BBa_J23118. As a
control
E. coli MG1655 were cultivated without the plasmid. RT-qPCR measurements were carried out in technical triplicates. Standard deviation is shown
Figure 1: Average relative quantification of RFP expression compared to the expression of the house-keeping gene GAP in E. coli
MG1655.
Cells contained the plasmid BBa_J61002 with different promoters: BBa_J23100, BBa_J23112 and BBa_J23118. As a
control
E. coli MG1655 were cultivated without the plasmid. RT-qPCR measurements were carried out in technical triplicates. Standard deviation is shown