Difference between revisions of "Part:BBa K1893009"
| Line 12: | Line 12: | ||
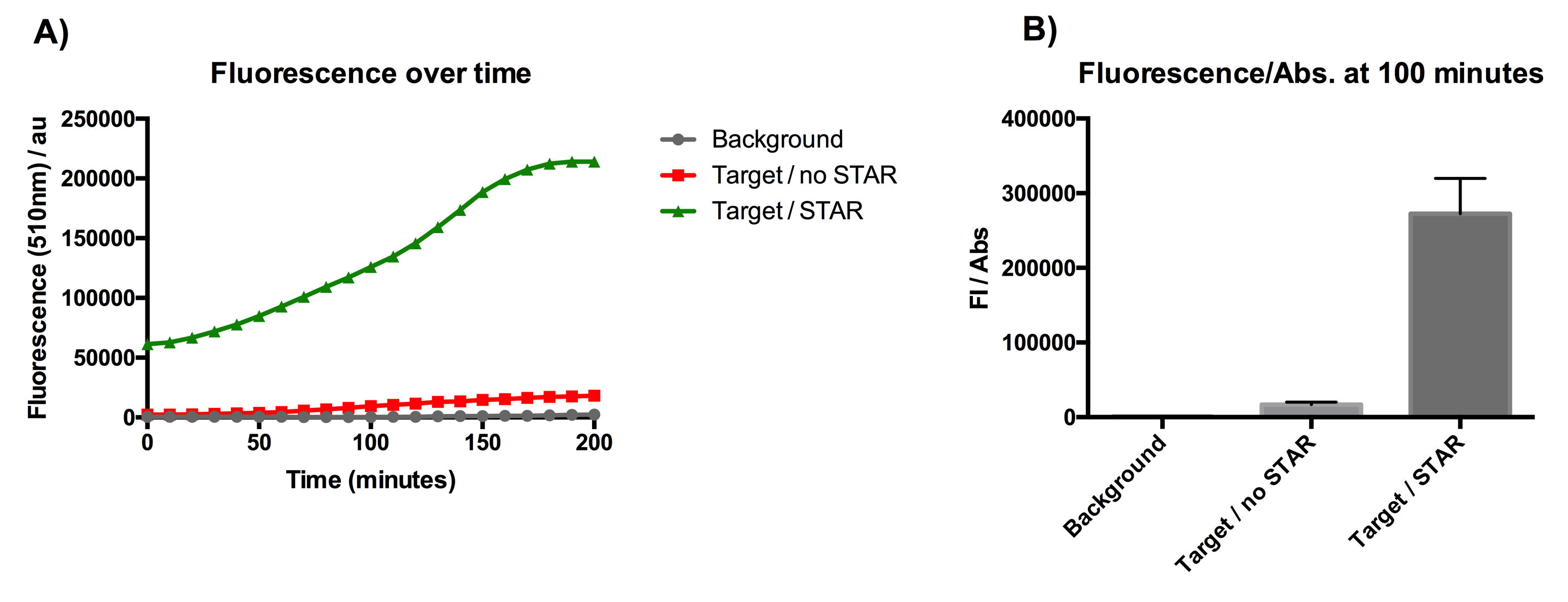
Figure 1: Characterisation of STAR system in TOP10 E. coli cells. (A) Normalised fluorescence monitored over time for cell lines incorporating the STAR system in the absence or presence of transcribed STAR molecules (B) Normalised endpoint fluorescence (100 minutes) for cell lines in the absence or presence of STAR molecules. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. Normalised fluorescence was calculated by dividing fluorescent signal by the O.D.600 value of the culture. Background was determined by the use of DH10B cells with no plasmid transformed. Error bars represent standard deviation from 3 technical repeats. | Figure 1: Characterisation of STAR system in TOP10 E. coli cells. (A) Normalised fluorescence monitored over time for cell lines incorporating the STAR system in the absence or presence of transcribed STAR molecules (B) Normalised endpoint fluorescence (100 minutes) for cell lines in the absence or presence of STAR molecules. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. Normalised fluorescence was calculated by dividing fluorescent signal by the O.D.600 value of the culture. Background was determined by the use of DH10B cells with no plasmid transformed. Error bars represent standard deviation from 3 technical repeats. | ||
| − | [[File:IC16_STAR2Agraph.png|300px|]][[File:IC16_STAR2B.png|300px|]][[File:IC16_STAR2C.png| | + | [[File:IC16_STAR2Agraph.png|300px|]][[File:IC16_STAR2B.png|300px|]][[File:IC16_STAR2C.png|345px|]] |
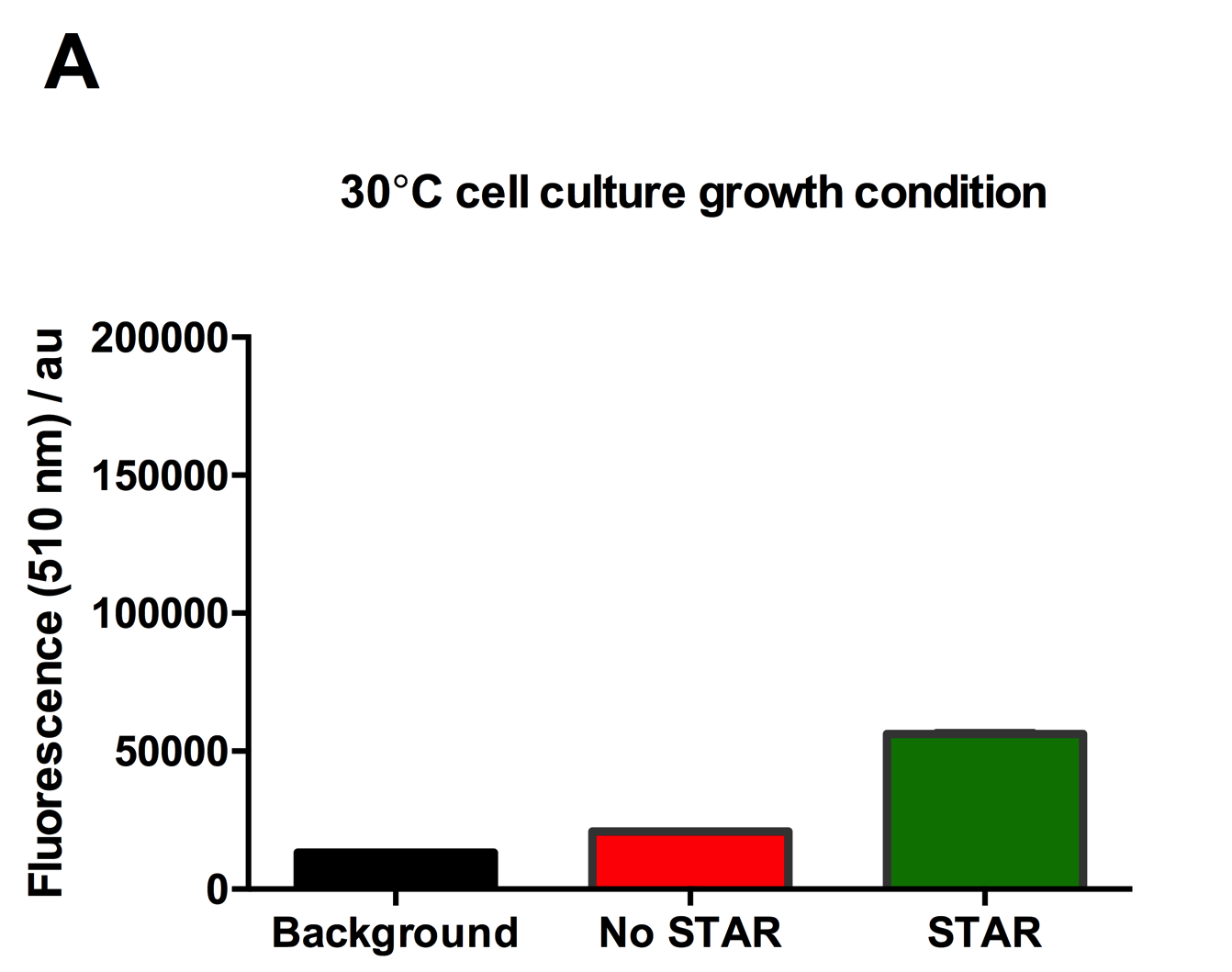
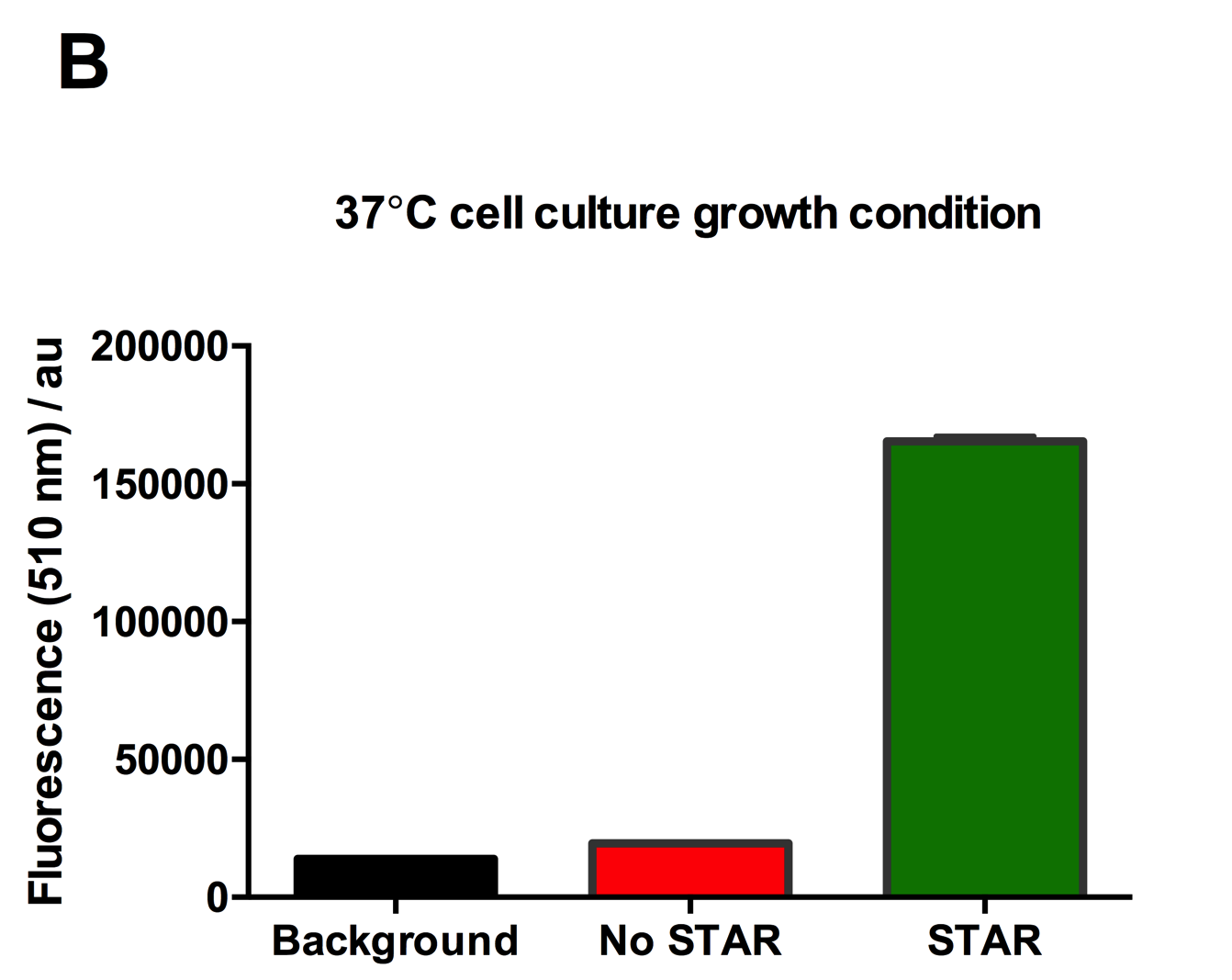
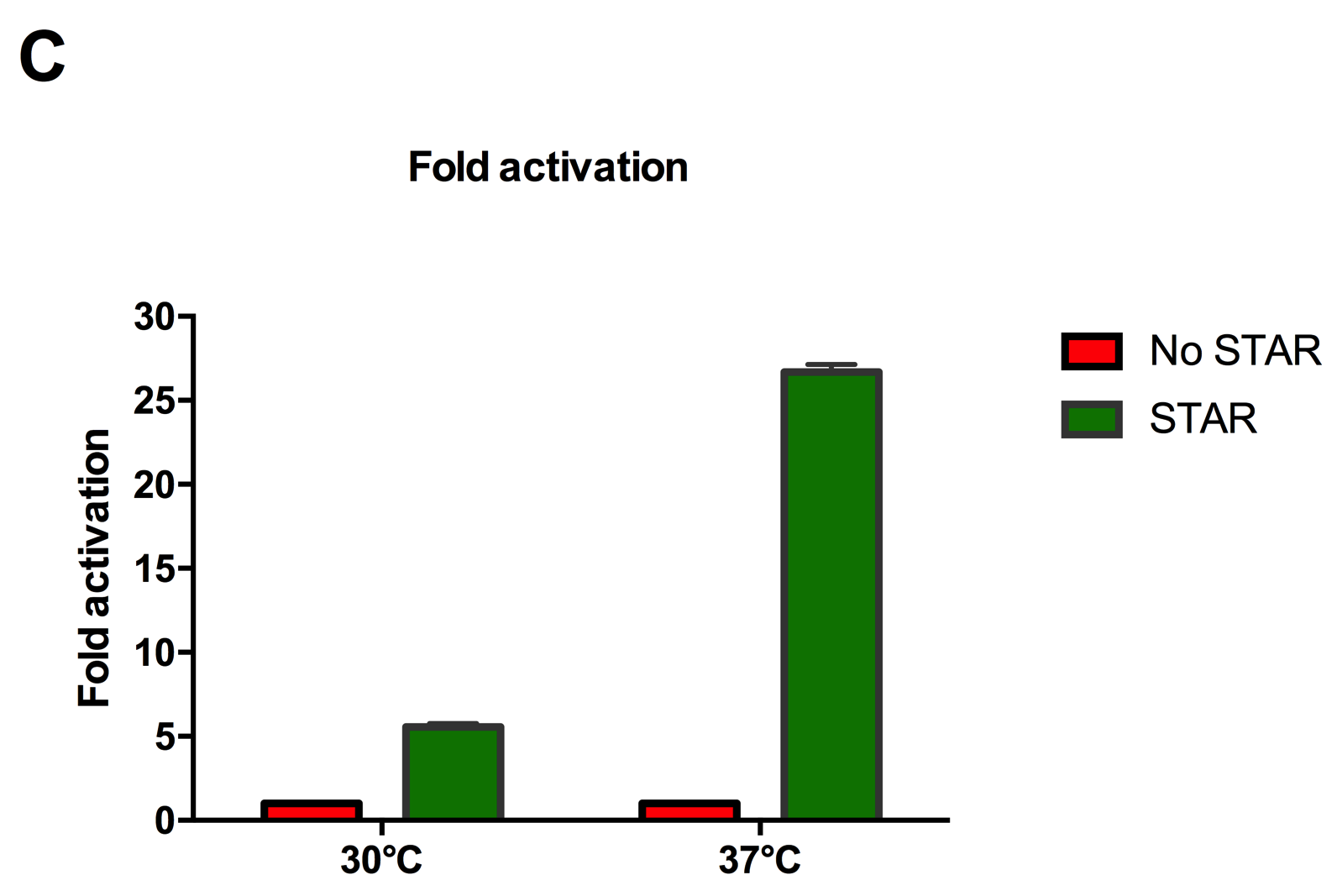
Figure 2: Characterisation of STAR system in TOP10 E. coli cells at different temperatures. (A) Cell culture fluorescence at 30°C (B) Cell culture fluorescence assay at 37°C. (C) Fold activation SFGFP expression in presence of STAR. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. The autofluorescence background control used is E. coli Top 10 cells with no reporter plasmid. Error bars represent standard deviation from 3 technical repeats. | Figure 2: Characterisation of STAR system in TOP10 E. coli cells at different temperatures. (A) Cell culture fluorescence at 30°C (B) Cell culture fluorescence assay at 37°C. (C) Fold activation SFGFP expression in presence of STAR. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. The autofluorescence background control used is E. coli Top 10 cells with no reporter plasmid. Error bars represent standard deviation from 3 technical repeats. | ||
Revision as of 15:57, 23 October 2016
STAR inducible superfolder GFP (pAD1.S5+SF-GFP)
This is the STAR repoter plasmid containing the pAD1.S5 attenuator sequence upstream to the reporter gene SFGFP. Transcription of SFGFP is also controlled by constitutive promoter j23119 and terminator TrrnB.
Usage and Biology
Figure 1: Characterisation of STAR system in TOP10 E. coli cells. (A) Normalised fluorescence monitored over time for cell lines incorporating the STAR system in the absence or presence of transcribed STAR molecules (B) Normalised endpoint fluorescence (100 minutes) for cell lines in the absence or presence of STAR molecules. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. Normalised fluorescence was calculated by dividing fluorescent signal by the O.D.600 value of the culture. Background was determined by the use of DH10B cells with no plasmid transformed. Error bars represent standard deviation from 3 technical repeats.
Figure 2: Characterisation of STAR system in TOP10 E. coli cells at different temperatures. (A) Cell culture fluorescence at 30°C (B) Cell culture fluorescence assay at 37°C. (C) Fold activation SFGFP expression in presence of STAR. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. The autofluorescence background control used is E. coli Top 10 cells with no reporter plasmid. Error bars represent standard deviation from 3 technical repeats.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 569
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 959
Illegal BsaI site found at 1137