Difference between revisions of "Part:BBa K1907000"
| Line 14: | Line 14: | ||
<p> | <p> | ||
<b>Previous use</b></p> | <b>Previous use</b></p> | ||
| − | The part has been expressed in <i>S. cerevisiae</i> strains W303α and SS328-leu, in the pRS415 plasmid under GPD1 and GAL1 promoters. During use, the part has not been used within the context of the BioBricks standard prefix and suffix; instead, the part has been directly flanked by SpeI and XhoI restriction sites (5’ and 3’, respectively), which were also used for cloning into the pRS415 plasmid. Expression results may vary when using the part in a different context; in particular, using the part in the context of the BioBricks prefix results in a T in the -3 position upstream of the start codon, which is strongly disfavored in eukaryotic transcription initiation (Kozak, 1996 | + | The part has been expressed in <i>S. cerevisiae</i> strains W303α and SS328-leu, in the pRS415 plasmid under GPD1 and GAL1 promoters. During use, the part has not been used within the context of the BioBricks standard prefix and suffix; instead, the part has been directly flanked by SpeI and XhoI restriction sites (5’ and 3’, respectively), which were also used for cloning into the pRS415 plasmid. Expression results may vary when using the part in a different context; in particular, using the part in the context of the BioBricks prefix results in a T in the -3 position upstream of the start codon, which is strongly disfavored in eukaryotic transcription initiation (Kozak, 1996; Cavener et al., 1991). |
| − | + | ||
| − | + | ||
<br> | <br> | ||
<p><b>Expression</b></p> | <p><b>Expression</b></p> | ||
Revision as of 01:23, 20 October 2016
Venus Yellow Fluorescent Protein
Introduction
Venus yellow fluorescent protein from Aequorea victoria (Nagai et al., 2002), codon-optimized for expression in Saccharomyces cerevisiae. The sequence is just the standalone protein, suitable for use as a fluorescent reporter. To use the part a fluorescent tag fused to another protein domain, it is recommended to add a linker.
Venus has an excitation wavelength of 515 nm and emission wavelength of 528 nm (Shaner et al., 2005). Venus has the advantage over many other YFPs of being less sensitive to environmental factors such as pH and chloride ions, and has a quicker rate of maturation (Nagai et al., 2002).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 644
Previous use
The part has been expressed in S. cerevisiae strains W303α and SS328-leu, in the pRS415 plasmid under GPD1 and GAL1 promoters. During use, the part has not been used within the context of the BioBricks standard prefix and suffix; instead, the part has been directly flanked by SpeI and XhoI restriction sites (5’ and 3’, respectively), which were also used for cloning into the pRS415 plasmid. Expression results may vary when using the part in a different context; in particular, using the part in the context of the BioBricks prefix results in a T in the -3 position upstream of the start codon, which is strongly disfavored in eukaryotic transcription initiation (Kozak, 1996; Cavener et al., 1991).
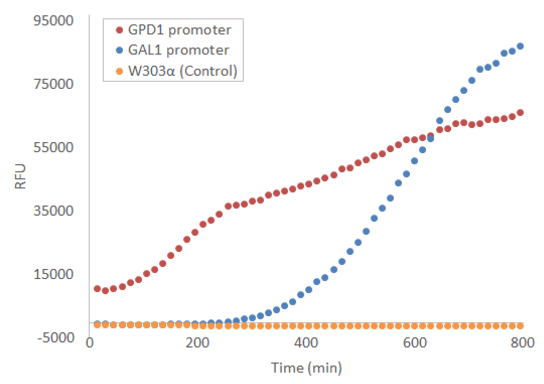
Expression
When used in the pRS415 plasmid, the produced fluorescence is not visible by eye, but can be easily measured with e.g. a microplate reader. Figure 1 presents the development of fluorescence under GPD1 and GAL1 promoters over time when a culture of starting OD=0.5 is grown at +30 °C with linear shaking of 731 cpm. Galactose induction is performed at t = 0.
References
Cavener, D.R., Ray, S.C, 1991. Eukaryotic start and stop translation sites. Nucleic Acids Research, 1991, 19(12), pp. 3185-3192
Kozak, M., 1996 Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell, 44(2), pp. 283-292
Nagai, T., Ibata, K., Park, E.S., Kubota, M., Mikoshiba, K., Atsushi, M., 2002, A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications, Nature Biotechnology, 20(1), pp. 87-90
Shaner, N.C., Steinbach, P.A. and Tsien, R.Y., 2005. A guide to choosing fluorescent proteins. Nature methods, 2(12), pp.905-909.