Difference between revisions of "Part:BBa K1933100"
(→RT-PCR) |
(→RT-PCR) |
||
| Line 23: | Line 23: | ||
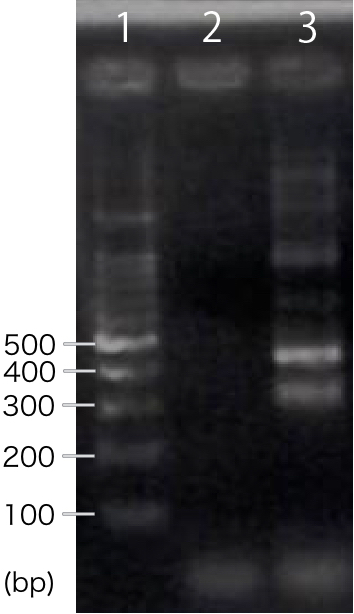
We designed PCR primers so that we would obtain PCR product with the length of 300 bp after RT-PCR. For negative control, reverse transcription was not performed. | We designed PCR primers so that we would obtain PCR product with the length of 300 bp after RT-PCR. For negative control, reverse transcription was not performed. | ||
[[file:T--Kyoto--RT-CBDcex.001.jpeg|200px|thumb|center|'''Fig.1''': ''' RT-PCR '''<br>1. marker, 2. INP-His-CBDcex (negative control), 3. INP-His-CBDcex]] | [[file:T--Kyoto--RT-CBDcex.001.jpeg|200px|thumb|center|'''Fig.1''': ''' RT-PCR '''<br>1. marker, 2. INP-His-CBDcex (negative control), 3. INP-His-CBDcex]] | ||
| − | + | A band corresponding to 300 bp was observed only in INP-His-scFv sample, not in negative control(Fig.1). This result suggests that INP-His-CBDcex was successfully transcribed in '' E.coli. '' | |
===Western blotting=== | ===Western blotting=== | ||
Revision as of 18:29, 16 October 2016
constitutive expression of CBDcex fused to INPNC with 6xHis tag
CBDcex fused to INPNC with 6xHis tag is one of a series of surface expressing fusion proteins that make up biodevice that aims to be therapeutic solution against norovirus infections. This protein in particular is a cellulose binding protein(CBDcex) fused to surface expression anchoring domain(INPNC), connected by a 6xHis tag to be easily identified by Western blotting. For more information, please visit [http://2016.igem.org/Team:Kyoto our wiki].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1012
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 132
Illegal NgoMIV site found at 465
Illegal AgeI site found at 883 - 1000COMPATIBLE WITH RFC[1000]
Usage
Characterization
colony PCR
RT-PCR
We designed PCR primers so that we would obtain PCR product with the length of 300 bp after RT-PCR. For negative control, reverse transcription was not performed.
A band corresponding to 300 bp was observed only in INP-His-scFv sample, not in negative control(Fig.1). This result suggests that INP-His-CBDcex was successfully transcribed in E.coli.
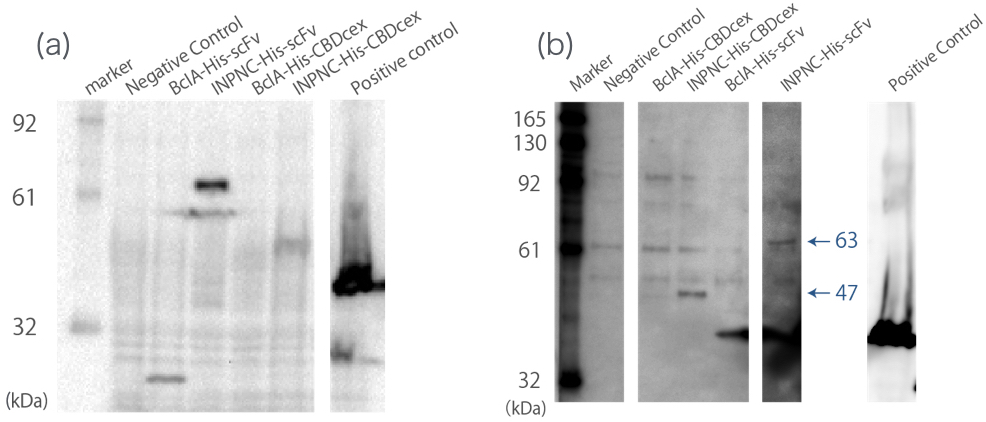
Western blotting
Reference
Judging
We fulfilled criteria listed below with this part.
- Validated Part/ Validated Contribution
- [http://2016.igem.org/Team:Kyoto/HP/Gold Integrated Human Practice]
- Improve a previous part or project
- [http://2016.igem.org/Team:Kyoto/Proof Proof of concept]