Difference between revisions of "Part:BBa R0071:Experience"
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | ===Improvement=== | + | ===Improvement and Characterize=== |
| − | + | Group: Tokyo Tech 2016 | |
| + | Author: Yoshio Takata | ||
| + | |||
| + | Summary of Improvement and Characterization: | ||
| + | |||
| + | I. Improved Prhl by iGEM 2014 Tokyo_Tech and characterize RhlR assay | ||
| + | |||
| + | II. Improvement of the wild type Prhl | ||
| + | |||
| + | III. Comparison of the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl | ||
| + | |||
| + | The sequences of these tow chosen mutants are shown in below. The small characters indicate the scar sequence and a single point mutation is colored with red. | ||
| + | |||
| + | Wild type Prhl (BBa_R0071) | ||
| + | TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTAAAAAGTGTTCtactagagAAAGAGGAGAAA | ||
| + | |||
| + | Prhl(NM) (BBa_K1949060) | ||
| + | TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTATAAAGTGTTCtactagagAAAGAGGAGAAA | ||
| + | |||
| + | Prhl(M) TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTATAAAGTGTTCtactagtaAAAGAGGAGAAA | ||
| + | |||
| + | III. Comparison the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl | ||
| + | Prhl(NM) was chosen from the many Prhl mutants, and by comparing Prhl(NM) to Prhl(LR), we obtained the result below (Fig.3-2-2-3-4). The reaction activity of Prhl(NM) to C4 was stronger than that of Prhl(LR), and Prhl(NM) did not react with C12 at all. | ||
| + | |||
| + | |||
| + | ====Discussion==== | ||
| + | |||
| + | Judging from the result of the experiment I, we found that Prhl(RL) activity was unexpectedly weak and enough expression could not be obtained without high C4 concentration which is designed by iGEM 2014 Tokyo_Tech. | ||
| + | |||
| + | The result that RhlR in the absence of C4 represses Prhl and the repression is contradicted when RhlR binds C4(1), RhlR without LVA tag repressed so strong that gfp could not express very much under the low C4 concentration (Fig.3-2-3-3-1). On the other hand, under the high C4 concentration, a lot of RhlR binded C4. Then the repression was contradicted and gfp expressed greatly(Fig.3-2-3-3-2). RhlR with LVA tag tended to degrade and the RhlR concentration in a cell was low. And that the repression by RhlR was not strong and gfp could express. RhlR without LVA tag could repress the growth rate of E. coli but the reason for this is unclear. | ||
| + | Prhl activity became stronger when the sequence of -10 region was changed from “TAAAAA” to "TATAAA", which is known as a part of the strongest promoter sequence in E. coli. Prhl(M) had accidental two mutations in the scar sequence and it is not suitable for a new part of BioBrick. | ||
| + | |||
| + | We think that the combination of the AHL regulators and the corresponding promoters also could control the expression level of the target gene. In our project, the combination of rhlR with LVA tag and Prhl(LR) might work well in our final genetic circuits. However, Prhl(LR) has lux box, and thus, the promoter also reacts with C12 (crosstalk) unintentionally. In our final circuits, we have to use C12 for Plux activation, and thus, Prhl(LR) does not suit for our final circuits. Then, we introduced a single point mutation into wild type Prhl (BBa_R0071), we obtained the Prhl(NM) of which activity was stronger than Prhl(LR). Comparing the Prhl(NM) to Prhl(LR), Prhl(NM) did not show crosstalking with C12 and the SN ratio of Prhl(NM) was higher than that of Prhl(LR). The SN ratio Prhl(LR) to C4 was small, because leaky expression from Prhl(LR) was strong. | ||
| + | |||
| + | As a conclusion, the combination of rhlR with LVA tag and Prhl(NM) is best for our final genetic circuits. | ||
| + | |||
| + | ====Materials and Methods==== | ||
| + | =====Construction===== | ||
| + | -Strain | ||
| + | All the plasmids were prepared in XL1-Blue strain. | ||
| + | |||
| + | -Plasmids | ||
| + | I. Improved Prhl by iGEM 2014 Tokyo_Tech and rhlR assay | ||
| + | A. Pcon-rbs-rhlR-LVA (BBa_C0071) (pSB6A1), Prhl(LR)(BBa_K1529310)-gfp (pSB3K3) (Fig. 3-2-3-5-1) | ||
| + | |||
| + | B. Pcon-rhlR(BBa_C0171) (pSB6A1), Prhl(LR)-gfp (pSB3K3) (Fig. 3-2-3-5-2) | ||
| + | C. Pcon-rhlR-LVA (pSB6A1), Prhl(RL)(BBa_K1529300)-gfp (pSB3K3) (Fig. 3-2-3-5-3) | ||
| + | D. Pcon-rhlR (pSB6A1), Prhl(RL)-gfp (pSB3K3) (Fig. 3-2-3-5-4) | ||
| + | E. Pcon-rhlR-LVA (pSB6A1), Pcon-gfp (pSB3K3) …Positive control (Fig. 3-2-3-5-5) | ||
| + | F. pSB6A1, pSB3K3 …Nagative control (Fig. 3-2-3-5-6) | ||
| + | |||
| + | II. Improvment the wild type Prhl | ||
| + | A. Prhl(BBa_R0071)-gfp (pSB3K3) (Fig. 3-2-3-5-7) | ||
| + | |||
| + | B. Pcon-rhlR-LVA (pSB6A1), Prhl(NM)(BBa_ K1949060 )-gfp (pSB3K3) (Fig. 3-2-3-5-8) | ||
| + | C. Pcon-rhlR-LVA (pSB6A1), Prhl(M)-gfp (pSB3K3) (Fig. 3-2-3-5-9) | ||
| + | D. Pcon-rhlR-LVA (pSB6A1), Pcon-gfp (pSB3K3) …Positive control (Fig. 3-2-3-5-10) | ||
| + | E. pSB6A1, pSB3K3 …Nagative control (Fig. 3-2-3-5-11) | ||
| + | |||
| + | III. Comparison the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl | ||
| + | A. Pcon-rhlR-LVA (pSB6A1), Prhl(LR)-gfp (pSB3K3) (Fig. 3-2-3-5-12) | ||
| + | B. Pcon-rhlR-LVA (pSB6A1), Prhl(NM)-gfp (pSB3K3) (Fig. 3-2-3-5-13) | ||
| + | C. Pcon-rhlR-LVA (pSB6A1), Pcon-gfp (pSB3K3) …Positive control (Fig. 3-2-3-5-14) | ||
| + | D. pSB6A1, pSB3K3…Negative control (Fig. 3-2-3-5-15) | ||
| + | |||
| + | -Medium | ||
| + | LB medium AK | ||
| + | LB medium containing ampicillin (50 microg/ mL) and kanamycin (50 microg/ mL) | ||
| + | LB medium K | ||
| + | LB medium containing kanamycin (50 microg/ mL) | ||
| + | |||
| + | |||
| + | =====Assay protocol===== | ||
| + | The following experiments is performed at 37℃ unless otherwise stated. | ||
| + | |||
| + | I. Improved Prhl by iGEM 2014 Tokyo_Tech and rhlR assay | ||
| + | |||
| + | 1) Prepare overnight culture for each sample in 3mL LB medium AK with vigorous shaking. | ||
| + | |||
| + | 2) Dilute the overnight cultures to 1 / 60 in fresh LB medium AK (1.2 mL). | ||
| + | |||
| + | 3) Incubate the fresh cultures for 1 h with vigorous shaking. | ||
| + | |||
| + | 4) Add C4 or DMSO to each 500 microL sample at the final concentration 10 microM or 100 microM. | ||
| + | |||
| + | 5) Incubate the samples for 4 h with vigorous shaking. | ||
| + | |||
| + | 6) Add 100 microL of the samples into each well of a plate reader. | ||
| + | |||
| + | 7) Measure the RFU intensity of GFP at 490 nm as an exciting wavelength, 525 nm as a measurement wavelength. | ||
| + | |||
| + | 8) Measure the Turbidity at 600 nm. | ||
| + | |||
| + | |||
| + | II. Improvement the wild type Prhl (BBa_R0071) | ||
| + | |||
| + | 1) Introduce a point mutation to -10 region of Prhl (BBa_R0071)-rbs-gfp (pSB3K3) by inverse PCR using xild type Prhl(BBa_R0071)-rbs-gfp as a template and divergent primers. | ||
| + | |||
| + | The sequences of the six forward premiers and of one reverse primer are shown below. | ||
| + | |||
| + | WT 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctaaaaagtgttctactagtagcg-3' | ||
| + | |||
| + | F-Mu1 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggntaaaaagtgttctactagtagcg-3' | ||
| + | |||
| + | F-Mu2 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggcnaaaaagtgttctactagtagcg-3' | ||
| + | |||
| + | F-Mu3 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctnaaaagtgttctactagtagcg-3' | ||
| + | |||
| + | F-Mu4 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctanaaagtgttctactagtagcg-3' | ||
| + | |||
| + | F-Mu5 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctaanaagtgttctactagtagcg-3' | ||
| + | |||
| + | F-Mu6 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctaaanagtgttctactagtagcg-3' | ||
| + | |||
| + | R 5'-aattcgaaagctaacggtaactgcca-3' (5’ phosphorylated) | ||
| + | |||
| + | 2) Prepare strains containing the pMu1~6 plasmids containing mutated Prhl-rbs-gfp(pSB3K3). | ||
| + | |||
| + | 3) Streak the transformants on an agar plate containing kanamycin (50 microg/ mL) and incubate overnight. | ||
| + | |||
| + | 4) Pore the LB medium K at the extent of covering the surface of the agar, and collect the colonies with a spreader. Then, incubate them in 5 mL LB medium K for 16 h with vigorous shaking. | ||
| + | |||
| + | 5) Extract the plasmid, and introduce Prhl(mut)-gfp (pSB3K3) and Pcon-rhlR-LVA to XL-1 Blue. | ||
| + | |||
| + | 6) Pore 200 microL LB medium AK containing C4 (6 microM) in one well of a 96-well plate and inoculate one colony to the well. Incubate the plate for 16 h. | ||
| + | |||
| + | 7) Transfer 100 microL of the culture from the above samples to another plate that is suitable for RFU measurement. Measure the RFU of GFP at 490 nm as an exciting wavelength and 525 nm as an emission wavelength as well as the turbidity at 600 nm. | ||
| + | |||
| + | 8) Select mutants that have higher SN ratio than wild type Prhl (BBa_R0071) and grow them in 3mL LB medium AK overnight with vigorous shaking. | ||
| + | |||
| + | 9) Dilute the overnight cultures to 1 / 60 in fresh LB medium AK (1.2 mL). | ||
| + | |||
| + | 10) Incubate the fresh cultures for 1 h with vigorous shaking. | ||
| + | |||
| + | 11) Add C4, C12 or DMSO into each 500 microL sample at the final concentration 10 microM. | ||
| + | |||
| + | 12) Incubate the samples for 4 h with vigorous shaking. | ||
| + | |||
| + | 13) Add 100 microL of the samples into each well of a plate reader. | ||
| + | |||
| + | 14) Measure the RFU of GFP at 490 nm as an exciting wavelength, 525 nm as a measurement wavelength. | ||
| + | |||
| + | 15) Measure the Turbidity at 600 nm. | ||
| + | |||
| + | |||
| + | |||
| + | III. Comparison the improved Prhl promoter by iGEM 2014 Tokyo_Tech to our original improved Prhl promoter | ||
| + | |||
| + | 1) Prepare overnight cultures for each sample in 3mL LB medium AK with vigorous shaking. | ||
| + | |||
| + | 2) Dilute the overnight cultures to 1 / 60 in fresh LB medium AK (1.2 mL). | ||
| + | |||
| + | 3) Incubate the fresh cultures for 1 h with vigorous shaking. | ||
| + | |||
| + | 4) Add C4, C12 or DMSO into each 500 microL sample at the final concentration 10 microM. | ||
| + | |||
| + | 5) Incubate the samples for 4 h with vigorous shaking. | ||
| + | |||
| + | 6) Add 100 microL of the samples into each well of a plate reader. | ||
| + | |||
| + | 7) Measure the RFU of GFP at 490 nm as an exciting wavelength, 525 nm as a measurement wavelength. | ||
| + | |||
| + | 8) Measure the Turbidity at 600 nm. | ||
| + | |||
| + | =====Reference===== | ||
| + | (1) Gerardo Medina et al. (2003) Mechanism of Pseudomonas aeruginosa RhlR Transcriptional Regulation of the rhlAB Promoter. | ||
| + | BACTERIOLOGY 185: 5976-5983 | ||
| + | |||
| + | (2) John S. Chuang et al. (2009) Simpson’s Paradox in a Synthetic Microbial System. SCIENCE 323: 272-275 | ||
| + | |||
| + | |||
| + | ===Improvement=== | ||
====iGEM 2014 Tokyo_Tech==== | ====iGEM 2014 Tokyo_Tech==== | ||
Revision as of 01:30, 15 October 2016
Improvement and Characterize
Group: Tokyo Tech 2016
Author: Yoshio Takata
Summary of Improvement and Characterization:
I. Improved Prhl by iGEM 2014 Tokyo_Tech and characterize RhlR assay
II. Improvement of the wild type Prhl
III. Comparison of the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl
The sequences of these tow chosen mutants are shown in below. The small characters indicate the scar sequence and a single point mutation is colored with red.
Wild type Prhl (BBa_R0071) TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTAAAAAGTGTTCtactagagAAAGAGGAGAAA
Prhl(NM) (BBa_K1949060) TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTATAAAGTGTTCtactagagAAAGAGGAGAAA
Prhl(M) TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTATAAAGTGTTCtactagtaAAAGAGGAGAAA
III. Comparison the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl Prhl(NM) was chosen from the many Prhl mutants, and by comparing Prhl(NM) to Prhl(LR), we obtained the result below (Fig.3-2-2-3-4). The reaction activity of Prhl(NM) to C4 was stronger than that of Prhl(LR), and Prhl(NM) did not react with C12 at all.
Discussion
Judging from the result of the experiment I, we found that Prhl(RL) activity was unexpectedly weak and enough expression could not be obtained without high C4 concentration which is designed by iGEM 2014 Tokyo_Tech.
The result that RhlR in the absence of C4 represses Prhl and the repression is contradicted when RhlR binds C4(1), RhlR without LVA tag repressed so strong that gfp could not express very much under the low C4 concentration (Fig.3-2-3-3-1). On the other hand, under the high C4 concentration, a lot of RhlR binded C4. Then the repression was contradicted and gfp expressed greatly(Fig.3-2-3-3-2). RhlR with LVA tag tended to degrade and the RhlR concentration in a cell was low. And that the repression by RhlR was not strong and gfp could express. RhlR without LVA tag could repress the growth rate of E. coli but the reason for this is unclear. Prhl activity became stronger when the sequence of -10 region was changed from “TAAAAA” to "TATAAA", which is known as a part of the strongest promoter sequence in E. coli. Prhl(M) had accidental two mutations in the scar sequence and it is not suitable for a new part of BioBrick.
We think that the combination of the AHL regulators and the corresponding promoters also could control the expression level of the target gene. In our project, the combination of rhlR with LVA tag and Prhl(LR) might work well in our final genetic circuits. However, Prhl(LR) has lux box, and thus, the promoter also reacts with C12 (crosstalk) unintentionally. In our final circuits, we have to use C12 for Plux activation, and thus, Prhl(LR) does not suit for our final circuits. Then, we introduced a single point mutation into wild type Prhl (BBa_R0071), we obtained the Prhl(NM) of which activity was stronger than Prhl(LR). Comparing the Prhl(NM) to Prhl(LR), Prhl(NM) did not show crosstalking with C12 and the SN ratio of Prhl(NM) was higher than that of Prhl(LR). The SN ratio Prhl(LR) to C4 was small, because leaky expression from Prhl(LR) was strong.
As a conclusion, the combination of rhlR with LVA tag and Prhl(NM) is best for our final genetic circuits.
Materials and Methods
Construction
-Strain All the plasmids were prepared in XL1-Blue strain.
-Plasmids I. Improved Prhl by iGEM 2014 Tokyo_Tech and rhlR assay A. Pcon-rbs-rhlR-LVA (BBa_C0071) (pSB6A1), Prhl(LR)(BBa_K1529310)-gfp (pSB3K3) (Fig. 3-2-3-5-1)
B. Pcon-rhlR(BBa_C0171) (pSB6A1), Prhl(LR)-gfp (pSB3K3) (Fig. 3-2-3-5-2) C. Pcon-rhlR-LVA (pSB6A1), Prhl(RL)(BBa_K1529300)-gfp (pSB3K3) (Fig. 3-2-3-5-3) D. Pcon-rhlR (pSB6A1), Prhl(RL)-gfp (pSB3K3) (Fig. 3-2-3-5-4) E. Pcon-rhlR-LVA (pSB6A1), Pcon-gfp (pSB3K3) …Positive control (Fig. 3-2-3-5-5) F. pSB6A1, pSB3K3 …Nagative control (Fig. 3-2-3-5-6)
II. Improvment the wild type Prhl A. Prhl(BBa_R0071)-gfp (pSB3K3) (Fig. 3-2-3-5-7)
B. Pcon-rhlR-LVA (pSB6A1), Prhl(NM)(BBa_ K1949060 )-gfp (pSB3K3) (Fig. 3-2-3-5-8) C. Pcon-rhlR-LVA (pSB6A1), Prhl(M)-gfp (pSB3K3) (Fig. 3-2-3-5-9) D. Pcon-rhlR-LVA (pSB6A1), Pcon-gfp (pSB3K3) …Positive control (Fig. 3-2-3-5-10) E. pSB6A1, pSB3K3 …Nagative control (Fig. 3-2-3-5-11)
III. Comparison the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl A. Pcon-rhlR-LVA (pSB6A1), Prhl(LR)-gfp (pSB3K3) (Fig. 3-2-3-5-12) B. Pcon-rhlR-LVA (pSB6A1), Prhl(NM)-gfp (pSB3K3) (Fig. 3-2-3-5-13) C. Pcon-rhlR-LVA (pSB6A1), Pcon-gfp (pSB3K3) …Positive control (Fig. 3-2-3-5-14) D. pSB6A1, pSB3K3…Negative control (Fig. 3-2-3-5-15)
-Medium LB medium AK LB medium containing ampicillin (50 microg/ mL) and kanamycin (50 microg/ mL) LB medium K LB medium containing kanamycin (50 microg/ mL)
Assay protocol
The following experiments is performed at 37℃ unless otherwise stated.
I. Improved Prhl by iGEM 2014 Tokyo_Tech and rhlR assay
1) Prepare overnight culture for each sample in 3mL LB medium AK with vigorous shaking.
2) Dilute the overnight cultures to 1 / 60 in fresh LB medium AK (1.2 mL).
3) Incubate the fresh cultures for 1 h with vigorous shaking.
4) Add C4 or DMSO to each 500 microL sample at the final concentration 10 microM or 100 microM.
5) Incubate the samples for 4 h with vigorous shaking.
6) Add 100 microL of the samples into each well of a plate reader.
7) Measure the RFU intensity of GFP at 490 nm as an exciting wavelength, 525 nm as a measurement wavelength.
8) Measure the Turbidity at 600 nm.
II. Improvement the wild type Prhl (BBa_R0071)
1) Introduce a point mutation to -10 region of Prhl (BBa_R0071)-rbs-gfp (pSB3K3) by inverse PCR using xild type Prhl(BBa_R0071)-rbs-gfp as a template and divergent primers.
The sequences of the six forward premiers and of one reverse primer are shown below.
WT 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctaaaaagtgttctactagtagcg-3'
F-Mu1 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggntaaaaagtgttctactagtagcg-3'
F-Mu2 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggcnaaaaagtgttctactagtagcg-3'
F-Mu3 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctnaaaagtgttctactagtagcg-3'
F-Mu4 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctanaaagtgttctactagtagcg-3'
F-Mu5 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctaanaagtgttctactagtagcg-3'
F-Mu6 5'-tcctgtgaaatctggcagttaccgttagctttcgaatt ggctaaanagtgttctactagtagcg-3'
R 5'-aattcgaaagctaacggtaactgcca-3' (5’ phosphorylated)
2) Prepare strains containing the pMu1~6 plasmids containing mutated Prhl-rbs-gfp(pSB3K3).
3) Streak the transformants on an agar plate containing kanamycin (50 microg/ mL) and incubate overnight.
4) Pore the LB medium K at the extent of covering the surface of the agar, and collect the colonies with a spreader. Then, incubate them in 5 mL LB medium K for 16 h with vigorous shaking.
5) Extract the plasmid, and introduce Prhl(mut)-gfp (pSB3K3) and Pcon-rhlR-LVA to XL-1 Blue.
6) Pore 200 microL LB medium AK containing C4 (6 microM) in one well of a 96-well plate and inoculate one colony to the well. Incubate the plate for 16 h.
7) Transfer 100 microL of the culture from the above samples to another plate that is suitable for RFU measurement. Measure the RFU of GFP at 490 nm as an exciting wavelength and 525 nm as an emission wavelength as well as the turbidity at 600 nm.
8) Select mutants that have higher SN ratio than wild type Prhl (BBa_R0071) and grow them in 3mL LB medium AK overnight with vigorous shaking.
9) Dilute the overnight cultures to 1 / 60 in fresh LB medium AK (1.2 mL).
10) Incubate the fresh cultures for 1 h with vigorous shaking.
11) Add C4, C12 or DMSO into each 500 microL sample at the final concentration 10 microM.
12) Incubate the samples for 4 h with vigorous shaking.
13) Add 100 microL of the samples into each well of a plate reader.
14) Measure the RFU of GFP at 490 nm as an exciting wavelength, 525 nm as a measurement wavelength.
15) Measure the Turbidity at 600 nm.
III. Comparison the improved Prhl promoter by iGEM 2014 Tokyo_Tech to our original improved Prhl promoter
1) Prepare overnight cultures for each sample in 3mL LB medium AK with vigorous shaking.
2) Dilute the overnight cultures to 1 / 60 in fresh LB medium AK (1.2 mL).
3) Incubate the fresh cultures for 1 h with vigorous shaking.
4) Add C4, C12 or DMSO into each 500 microL sample at the final concentration 10 microM.
5) Incubate the samples for 4 h with vigorous shaking.
6) Add 100 microL of the samples into each well of a plate reader.
7) Measure the RFU of GFP at 490 nm as an exciting wavelength, 525 nm as a measurement wavelength.
8) Measure the Turbidity at 600 nm.
Reference
(1) Gerardo Medina et al. (2003) Mechanism of Pseudomonas aeruginosa RhlR Transcriptional Regulation of the rhlAB Promoter. BACTERIOLOGY 185: 5976-5983
(2) John S. Chuang et al. (2009) Simpson’s Paradox in a Synthetic Microbial System. SCIENCE 323: 272-275
Improvement
iGEM 2014 Tokyo_Tech
We, Tokyo Tech 2014, improved Prhl Promoter (BBa_R0071) by changing LuxR binding site of Plux Promoter to RhlR binding site.
We dsigned new Prhl promoter
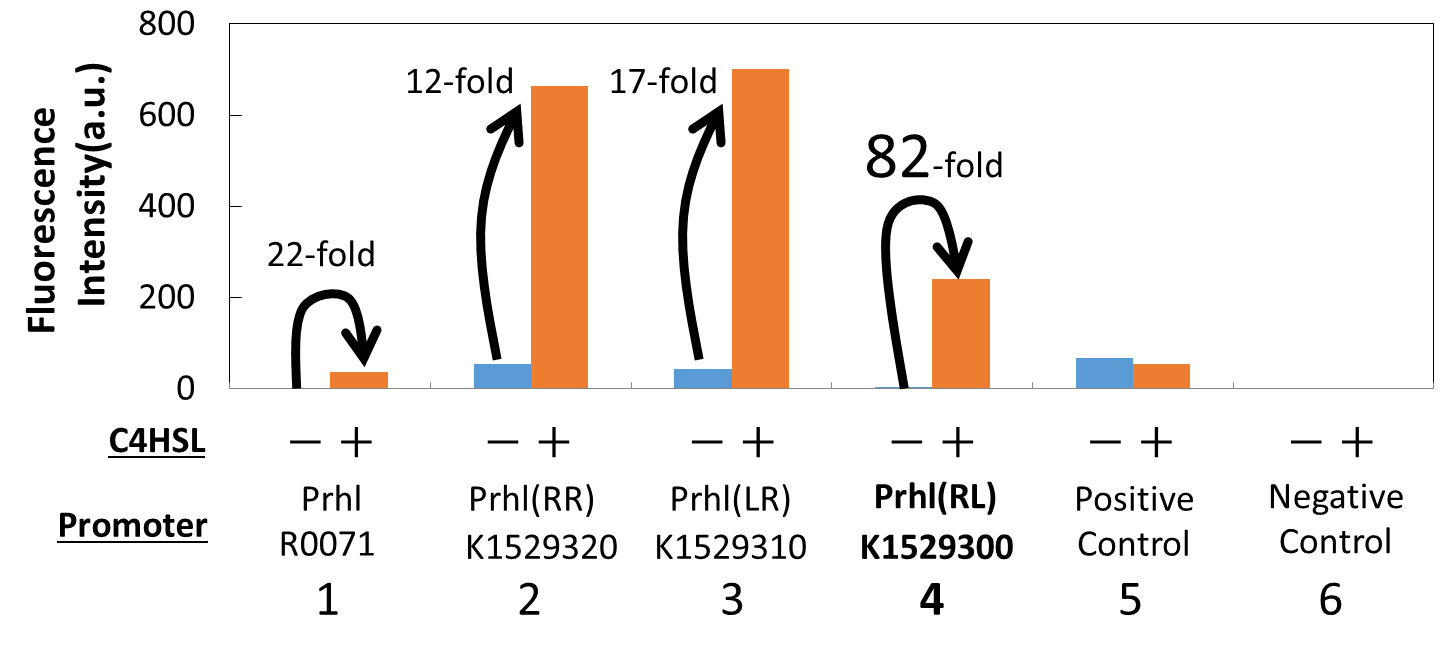
We measured the GFP expression with the four different promoters (Prhl : BBa_R0071, Prhl(RR) : BBa_K1529320, Prhl(LR) : BBa_K1529310 and Prhl(RL) : BBa_K1529300) by flow cytometer. Each promoter was tested in the presence and also in the absence of C4HSL.
Fig. 1 shows the fluorescence intensity detected by flow cytometer.
From the result, when induced by C4HSL, Prhl(RR) promoter showed higher maximum expression level and higher leak than the original Prhl promoter (BBa_R0071).
Although Prhl(RL) promoter had lower maximum expression level compared to Prhl(RR) promoter, it had the highest induced/not-induced ratio.
This means Prhl(RL) promoter has little leak.
Therefore, we can say that Prhl(RL):BBa_K1529300 promoter is the best improved Prhl promoter due to the advantages of less leak and higher expression level.
For more information, see [http://2014.igem.org/Team:Tokyo_Tech/Experiment/Prhl_reporter_assay our work in Tokyo_Tech 2014 wiki].
Applications of BBa_R0071
User Reviews
UNIQ9ed9eae3ade742b4-partinfo-00000005-QINU UNIQ9ed9eae3ade742b4-partinfo-00000006-QINU
|
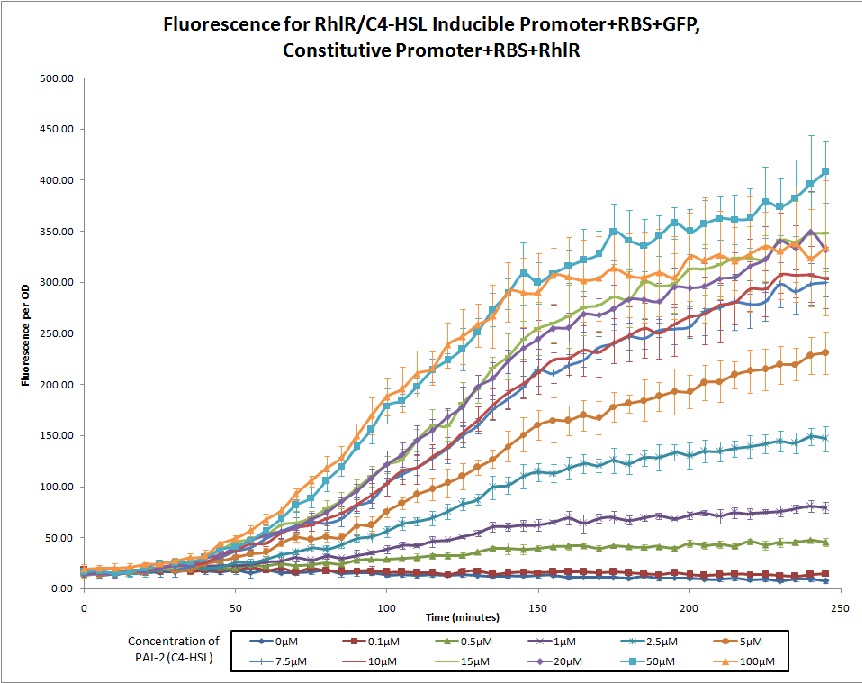
No review score entered. Northwestern 2011 |
The 2011 Northwestern iGEM team used this promoter as a unit within our Pseudomonas Aeruginosa biosensor. When this RhlR/C4-HSL regulated promoter is induced at varying concentrations of C4-HSL, we observed GFP fluorescence in accordance to the graph below. |