Difference between revisions of "Part:BBa K1694025"
ChinzLin00 (Talk | contribs) |
ChinzLin00 (Talk | contribs) |
||
| Line 13: | Line 13: | ||
'''1.Cloning''' | '''1.Cloning''' | ||
<br> | <br> | ||
| − | After assembling the DNA sequences from the basic parts, we recombined each Pcons+RBS+<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1694002">Lpp-OmpA-N</a>+ScFv gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each parts. The DNA sequence length of these parts is around 1100~1300 bp. In this PCR experiment, the PCR products' size should be near at 1300~1500 bp. The Fig. 3 showed the correct size of this part, and proved that we successfully ligated the sequence onto an ideal backbone. | + | After assembling the DNA sequences from the basic parts, we recombined each Pcons+RBS+<html><a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1694002">Lpp-OmpA-N</a></html>+ScFv gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each parts. The DNA sequence length of these parts is around 1100~1300 bp. In this PCR experiment, the PCR products' size should be near at 1300~1500 bp. The Fig. 3 showed the correct size of this part, and proved that we successfully ligated the sequence onto an ideal backbone. |
<div style="display: block;height: 500pt;"> | <div style="display: block;height: 500pt;"> | ||
[[File:PROHPCR.png|200px|thumb|left|'''Fig.3''' The PCR result of the Pcons+B0034+Lpp-OmpA-N+ScFv. The DNA sequence length is around 1100~1300 bp, so the PCR products should appear at 1300~1500 bp.]] | [[File:PROHPCR.png|200px|thumb|left|'''Fig.3''' The PCR result of the Pcons+B0034+Lpp-OmpA-N+ScFv. The DNA sequence length is around 1100~1300 bp, so the PCR products should appear at 1300~1500 bp.]] | ||
| Line 21: | Line 21: | ||
'''2.Cell staining experiment'''<br> | '''2.Cell staining experiment'''<br> | ||
| − | After cloning the part of <a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1694005">anti-HER2</a>, we were able to co-transform anti-HER2 with different fluorescence protein into our ''E. coli''. <br> | + | After cloning the part of <html><a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1694005">anti-HER2</a></html>, we were able to co-transform anti-HER2 with different fluorescence protein into our ''E. coli''. <br> |
The next step was to prove that our co-transformed product have successfully displayed ScFv of anti-HER2 and expressed fluorescence protein. | The next step was to prove that our co-transformed product have successfully displayed ScFv of anti-HER2 and expressed fluorescence protein. | ||
<br> | <br> | ||
Revision as of 07:11, 22 September 2015
Pcons+B0034+Lpp-OmpA-N+scFv(Anti-HER2)
Introduction:
This year we want to supply a customized platform. We provide two libraries of Pcon+RBS+OmpA-scFv and Pcons+RBS+Fluorescence+Ter for customers. Therefore, customers can choose any scFv and fluorescence proteins combination whatever they want. Our team will then co-transform the two plasmids into E. coli, which helps us tailor our product to the wishes of our customers.
Experiment
1.Cloning
After assembling the DNA sequences from the basic parts, we recombined each Pcons+RBS+Lpp-OmpA-N+ScFv gene to pSB1C3 backbones and conducted a PCR experiment to check the size of each parts. The DNA sequence length of these parts is around 1100~1300 bp. In this PCR experiment, the PCR products' size should be near at 1300~1500 bp. The Fig. 3 showed the correct size of this part, and proved that we successfully ligated the sequence onto an ideal backbone.
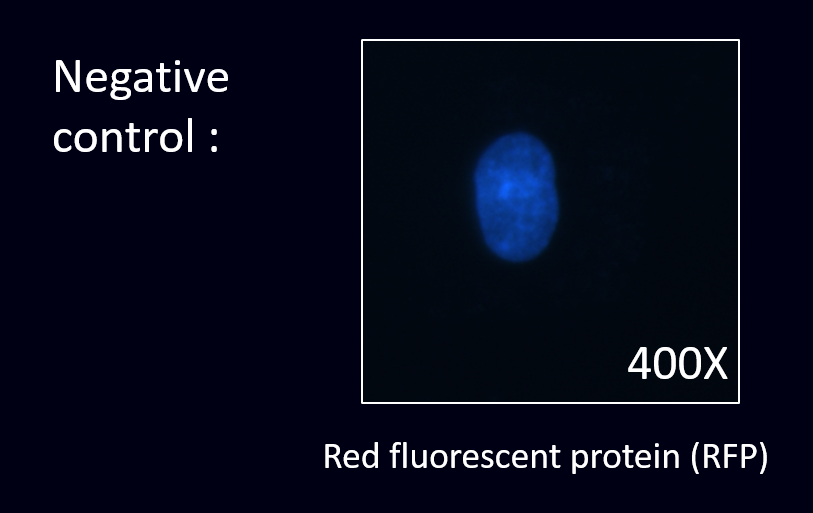
2.Cell staining experiment
After cloning the part of anti-HER2, we were able to co-transform anti-HER2 with different fluorescence protein into our E. coli.
The next step was to prove that our co-transformed product have successfully displayed ScFv of anti-HER2 and expressed fluorescence protein.
To prove this, we conducted the cell staining experiment by using the co-transformed E. coli to detect the HER2 in the cancer cell line.
Modeling:
In the modeling part, we discover optimum protein production time by using the genetic algorithm in Matlab.
We want to characterize the actual kinetics of this Hill-function based model that accurately reflects protein production time.
When we have the simulated protein production rate, the graph of protein production versus time can be drawn (Fig.1) (Fig.2) (Fig.3). Thus, we'll know the time of optimum production and the simulated one are fitted or not.
Co-transform



Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 741
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 451
- 1000COMPATIBLE WITH RFC[1000]