Difference between revisions of "Part:BBa K1692023"
Djcamenares (Talk | contribs) |
|||
| Line 5: | Line 5: | ||
Our goal was to create an autolysis sytem that can lyse cell after cell growth reaches a threshold density. To do so, we incorporated two previous BioBricks ptet ([https://parts.igem.org/Part:BBa_R0040 BBa_R0040]) and luxl generator ([https://parts.igem.org/Part:BBa_K805016 BBa_K805016]). We wanted luxl to be produced constantly; however, it should not be overexpressed so we use a moderate strength promoter, ptet. We sequenced the construct and showed that it matched the intended construct. Using ptet-luxl generator, we were successful in designing our autolysis system ([https://parts.igem.org/Part:BBa_K1692024 BBa_K1692024]). | Our goal was to create an autolysis sytem that can lyse cell after cell growth reaches a threshold density. To do so, we incorporated two previous BioBricks ptet ([https://parts.igem.org/Part:BBa_R0040 BBa_R0040]) and luxl generator ([https://parts.igem.org/Part:BBa_K805016 BBa_K805016]). We wanted luxl to be produced constantly; however, it should not be overexpressed so we use a moderate strength promoter, ptet. We sequenced the construct and showed that it matched the intended construct. Using ptet-luxl generator, we were successful in designing our autolysis system ([https://parts.igem.org/Part:BBa_K1692024 BBa_K1692024]). | ||
| − | + | ===Measuring Expression: Alma 2019=== | |
| − | + | ||
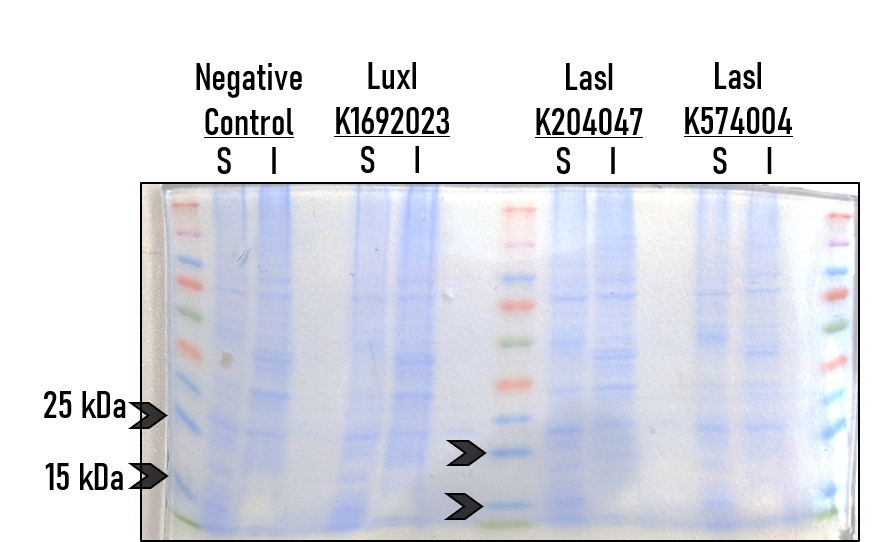
| + | To test the expression and activity of [https://parts.igem.org/Part:BBa_K204047 BBa_K204047], [https://parts.igem.org/Part:BBa_K574004 K574004], and K1692023, we (the Alma College iGEM team) took the following steps. Equal volumes of mid-log cells were resuspended in fresh LB + Chloramphenicol. These cultures were incubated for 6 hours at 37C in a tube rotator. Cells were pelleted by centrifugation - the supernatant was saved for detection of AHL molecules (all those synthesized during the incubation). | ||
| + | |||
| + | From the remaining cell pellet, proteins were isolated using the B-PER reagent (Thermo-Scientific). Soluble and Insoluble proteins were then run on an SDS PAGE gel. The gel was stained with coomaise, and density analsyis was performed with ImageJ. | ||
| + | |||
| + | [[File:T--Alma--AHLsynthaseExpression1.png]] | ||
| + | |||
| + | This is a representative image from several independent replicates. S = Soluble protein fraction, I = Insoluble protein fraction. The negative control used in this particular replicate was an uninduced culture of strain BBa_T9002. | ||
| + | |||
| + | [[File:T--Alma--AHLsynthaseGelDensity1.png]] | ||
| + | |||
| + | This is a density analysis of the soluble lanes in the previous gel image; in particular, for the region between the 35 kDa marker and 15 kDa marker. (Blue line, left to right). The lines show a normalized density for each protein sample, and highlight that there are no significant peaks for the AHL synthase BioBrick cultures above background in the 20-25 kDa range, which is where the AHL synthases would be expected to be found. | ||
| + | |||
| + | From these results we were barely able to detect any expression of the AHL synthase genes - proteins such as LuxI and LasI should be present as bands slightly below the 25 kDa marker. However, the supernatants from these same cultures did display AHL when tested with the biosensor strain [https://parts.igem.org/Part:BBa_T9002 BBa_T9002]. Thus, we conclude that the proteins are present but at very low quantities - likely a result of the degrons present, leading to a very short half-life. | ||
| + | |||
| + | At the same time as performing these experiments, we were able to detect protein expression for reporters such as RFP (BBa_J04450) and GFP (BBa_K515005), suggesting that our technique and experimental setup was not responsible for our inability to see expression of the AHL synthease genes. | ||
<!-- --> | <!-- --> | ||
Revision as of 00:56, 9 October 2019
Ptet + Luxl
Overview
Our goal was to create an autolysis sytem that can lyse cell after cell growth reaches a threshold density. To do so, we incorporated two previous BioBricks ptet (BBa_R0040) and luxl generator (BBa_K805016). We wanted luxl to be produced constantly; however, it should not be overexpressed so we use a moderate strength promoter, ptet. We sequenced the construct and showed that it matched the intended construct. Using ptet-luxl generator, we were successful in designing our autolysis system (BBa_K1692024).
Measuring Expression: Alma 2019
To test the expression and activity of BBa_K204047, K574004, and K1692023, we (the Alma College iGEM team) took the following steps. Equal volumes of mid-log cells were resuspended in fresh LB + Chloramphenicol. These cultures were incubated for 6 hours at 37C in a tube rotator. Cells were pelleted by centrifugation - the supernatant was saved for detection of AHL molecules (all those synthesized during the incubation).
From the remaining cell pellet, proteins were isolated using the B-PER reagent (Thermo-Scientific). Soluble and Insoluble proteins were then run on an SDS PAGE gel. The gel was stained with coomaise, and density analsyis was performed with ImageJ.
This is a representative image from several independent replicates. S = Soluble protein fraction, I = Insoluble protein fraction. The negative control used in this particular replicate was an uninduced culture of strain BBa_T9002.
This is a density analysis of the soluble lanes in the previous gel image; in particular, for the region between the 35 kDa marker and 15 kDa marker. (Blue line, left to right). The lines show a normalized density for each protein sample, and highlight that there are no significant peaks for the AHL synthase BioBrick cultures above background in the 20-25 kDa range, which is where the AHL synthases would be expected to be found.
From these results we were barely able to detect any expression of the AHL synthase genes - proteins such as LuxI and LasI should be present as bands slightly below the 25 kDa marker. However, the supernatants from these same cultures did display AHL when tested with the biosensor strain BBa_T9002. Thus, we conclude that the proteins are present but at very low quantities - likely a result of the degrons present, leading to a very short half-life.
At the same time as performing these experiments, we were able to detect protein expression for reporters such as RFP (BBa_J04450) and GFP (BBa_K515005), suggesting that our technique and experimental setup was not responsible for our inability to see expression of the AHL synthease genes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 717
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]