Difference between revisions of "Part:BBa K1399001"
m |
Carolinarp (Talk | contribs) |
||
| Line 13: | Line 13: | ||
[2] Andersen, J. B. et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–6 (1998). | [2] Andersen, J. B. et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–6 (1998). | ||
[3] Purcell, O., Grierson, C. S., Bernardo, M. Di & Savery, N. J. Temperature dependence of ssrA-tag mediated protein degradation. J. Biol. Eng. 6, 10 (2012). | [3] Purcell, O., Grierson, C. S., Bernardo, M. Di & Savery, N. J. Temperature dependence of ssrA-tag mediated protein degradation. J. Biol. Eng. 6, 10 (2012). | ||
| + | |||
| + | |||
| + | '''Contribution''' | ||
| + | Team: Valencia_UPV iGEM 2018 | ||
| + | Summarize: we have created part BBa_K2656024. It is the Monomeric red fluorescent protein with LVA degradation tag [https://parts.igem.org/Part:BBa_K1399001 BBa_K1399001] adapted into the Golden Gate assembly method. Thus, it is a basic part compatible with both Biobrick and [http://2018.igem.org/Team:Valencia_UPV/Design GoldenBraid 3.0] grammar. | ||
<partinfo>BBa_K1399001 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1399001 SequenceAndFeatures</partinfo> | ||
Revision as of 10:22, 17 October 2018
RFP from Discosoma striata (coral) with LVA-ssrA degradation tag
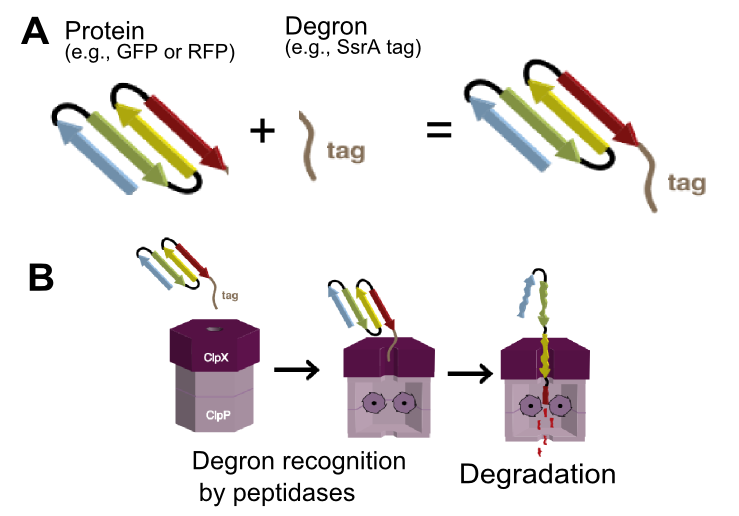
Mutant RFP from Discosoma striata (coral) (Part:BBa_E1010) with added LVA-ssrA degradation tag. The tag increases RFP turn-over rate, thus providing better temporal resolution of red fluorescence. In the same time, maximal fluorescence amplitudes will be lower as newly formed protein is degraded as soon as it is formed. LVA tag is commonly found attached to repressors in various gene networks (e.g. oscillators). The tag encodes peptide sequence AANDENYALVA and is recognized by ClpA and ClpX unfoldases and ClpX mediator SspB.[1] ClpA and ClpX then form a proteosome-like complex with ClpP protease and the protein is degraded (Figure 1).[1]
The final three residues of the tag determines the strength of interaction with ClpX and thus the final protein degradation rate.[2] The LVA tag is reported to lead to fast protein degradation, degrading GFP with rate -0.018 per minute.[2] However, be aware that exact protein degradation rate depends on multiple factors: ClpXP and ClpAP protease and SspB mediator concentrations, protein stability, Km of binding to the protease, temperature [3].
References: [1] Flynn, J. M. et al. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98, 10584–9 (2001). [2] Andersen, J. B. et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–6 (1998). [3] Purcell, O., Grierson, C. S., Bernardo, M. Di & Savery, N. J. Temperature dependence of ssrA-tag mediated protein degradation. J. Biol. Eng. 6, 10 (2012).
Contribution
Team: Valencia_UPV iGEM 2018
Summarize: we have created part BBa_K2656024. It is the Monomeric red fluorescent protein with LVA degradation tag BBa_K1399001 adapted into the Golden Gate assembly method. Thus, it is a basic part compatible with both Biobrick and [http://2018.igem.org/Team:Valencia_UPV/Design GoldenBraid 3.0] grammar.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 555
Illegal AgeI site found at 667 - 1000COMPATIBLE WITH RFC[1000]