Difference between revisions of "Part:BBa K1467101:Experience"
LedaCoelewij (Talk | contribs) (Further Characterization NRP-UEA) |
|||
| Line 8: | Line 8: | ||
iGEM14_NRP-UEA-Norwich have used this part in constructs to constitutively express proteins of interest, often providing a control to test the expression of each of our reporters. This allowed us to compare expression levels in contrast to when using promoters induced by external stimuli (BS3, PDF1.2, PR1), allowing us to select the most effective inducible promoters. | iGEM14_NRP-UEA-Norwich have used this part in constructs to constitutively express proteins of interest, often providing a control to test the expression of each of our reporters. This allowed us to compare expression levels in contrast to when using promoters induced by external stimuli (BS3, PDF1.2, PR1), allowing us to select the most effective inducible promoters. | ||
| + | |||
| + | |||
| + | <u> NRP-UEA 2015 </u> | ||
| + | |||
| + | At NRP-UEA, we aimed to produce a prebiotic to prevent colon cancer. To do this, we attempted to acylate/butrylate starch in plants. Methods to chemically acylate starch purified from plants already exist, but as they use strong chemicals and require heating they are not environmentally friendly. Using various acyltransferases, we hope to acylate starch in plants. We’ll be using a model plant, Nicotinia benthamiana, for initial tests because we can get results within a few days. We used Golden Gate Cloning and the Plant Standard Syntax described in RFC 106 and Patron et al (2015) to make our constructs. | ||
| + | |||
| + | All of our constructs contained the 35s Constitutive Promoter. This was done to drive the constitutive expression of our parts in the plants. We also added a chloroplast transit peptide, acyl transferase, and a 35s terminator to our constructs (BBa_K1618033‐036). In a second set of parts, we added a fluorescent tag, to confirm the function of our chloroplast transit peptide (BBa_K1618029‐032). | ||
| + | |||
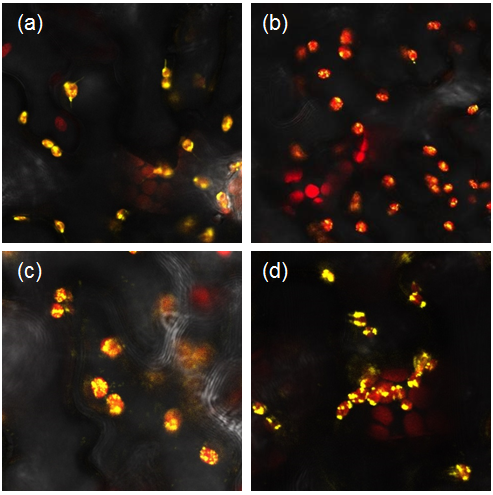
| + | We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves. | ||
| + | |||
| + | |||
| + | [[File:Confocal_Image_029-032.png]] | ||
| + | |||
| + | <i> Figure 1: Constructs BBa_K1618029‐032 contain a yellow fluorescent protein, as well as a chloroplast transit peptide. These are confocal microscopy images of the constructs infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from constructs (a) BBa_K1618029, (b) BBa_K1618031, (c) BBa_K1618032, and (d) BBa_K1618030. </i> | ||
| + | |||
| + | The above results not only indicate that our chloroplast transit peptide works, but also suggest that the 35s promoter that we used is GoldenGate compatible and functioning. | ||
| + | |||
===User Reviews=== | ===User Reviews=== | ||
Revision as of 11:57, 17 September 2015
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1467101
NRP-UEA 2014
iGEM14_NRP-UEA-Norwich have used this part in constructs to constitutively express proteins of interest, often providing a control to test the expression of each of our reporters. This allowed us to compare expression levels in contrast to when using promoters induced by external stimuli (BS3, PDF1.2, PR1), allowing us to select the most effective inducible promoters.
NRP-UEA 2015
At NRP-UEA, we aimed to produce a prebiotic to prevent colon cancer. To do this, we attempted to acylate/butrylate starch in plants. Methods to chemically acylate starch purified from plants already exist, but as they use strong chemicals and require heating they are not environmentally friendly. Using various acyltransferases, we hope to acylate starch in plants. We’ll be using a model plant, Nicotinia benthamiana, for initial tests because we can get results within a few days. We used Golden Gate Cloning and the Plant Standard Syntax described in RFC 106 and Patron et al (2015) to make our constructs.
All of our constructs contained the 35s Constitutive Promoter. This was done to drive the constitutive expression of our parts in the plants. We also added a chloroplast transit peptide, acyl transferase, and a 35s terminator to our constructs (BBa_K1618033‐036). In a second set of parts, we added a fluorescent tag, to confirm the function of our chloroplast transit peptide (BBa_K1618029‐032).
We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves.
Figure 1: Constructs BBa_K1618029‐032 contain a yellow fluorescent protein, as well as a chloroplast transit peptide. These are confocal microscopy images of the constructs infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from constructs (a) BBa_K1618029, (b) BBa_K1618031, (c) BBa_K1618032, and (d) BBa_K1618030.
The above results not only indicate that our chloroplast transit peptide works, but also suggest that the 35s promoter that we used is GoldenGate compatible and functioning.
User Reviews
UNIQ90763b78bc393955-partinfo-00000000-QINU UNIQ90763b78bc393955-partinfo-00000001-QINU