Difference between revisions of "Part:BBa K1033923"
| Line 10: | Line 10: | ||
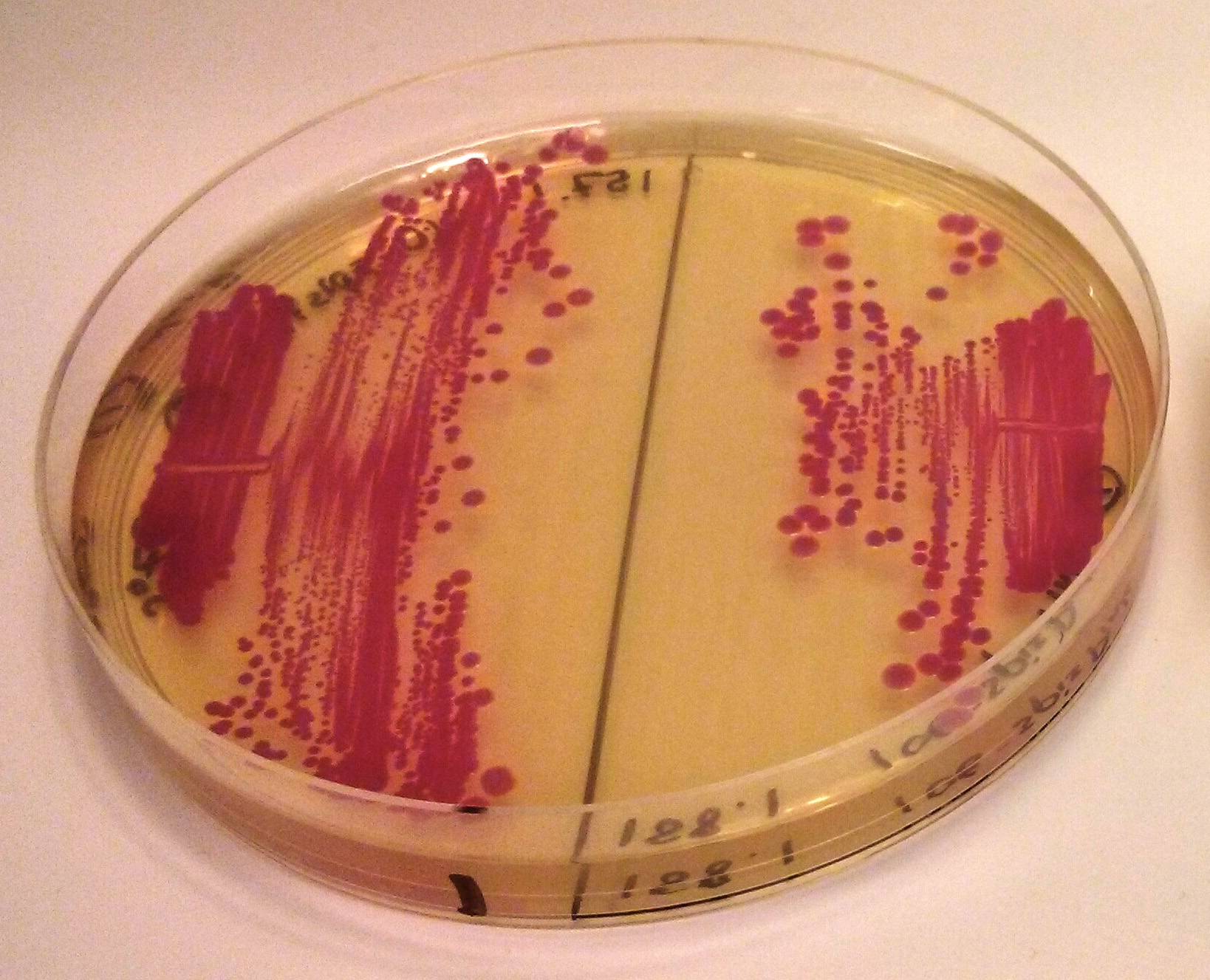
'''iGEM2013 Uppsala:''' The images above show ''E coli'' constitutively expressing spisPink <partinfo>BBa_K1033925</partinfo> from the high copy plasmid pSB1C3 from the promoters J23110 (left) and J23106 (right). | '''iGEM2013 Uppsala:''' The images above show ''E coli'' constitutively expressing spisPink <partinfo>BBa_K1033925</partinfo> from the high copy plasmid pSB1C3 from the promoters J23110 (left) and J23106 (right). | ||
| − | + | ===Characterisation=== | |
| + | [[File:T—Humboldt_Berlin—CharacteriastionSpisPinkAbs]] | ||
===Source=== | ===Source=== | ||
''Stylophora pistillata''. The protein was first extracted and characterized by Alieva et. al. under the name spisCP (GenBank: ABB17971.1). This version is codon optimized for '' E coli'' by Genscript. | ''Stylophora pistillata''. The protein was first extracted and characterized by Alieva et. al. under the name spisCP (GenBank: ABB17971.1). This version is codon optimized for '' E coli'' by Genscript. | ||
Revision as of 15:04, 13 October 2019
spisPink, pink chromoprotein (incl RBS, J23110)
This chromoprotein from the coral Stylophora pistillata, spisPink (also known as spisCP), naturally exhibits strong color when expressed. The protein has an absorption maximum at 560 nm giving it a pink color visible to the naked eye. The strong color is readily observed in both LB or on agar plates after less than 24 hours of incubation. The protein spisPink has significant sequence homologies with proteins in the GFP family.
Usage and Biology
This part is useful as a reporter.
iGEM2013 Uppsala: The images above show E coli constitutively expressing spisPink BBa_K1033925 from the high copy plasmid pSB1C3 from the promoters J23110 (left) and J23106 (right).
Characterisation
File:T—Humboldt Berlin—CharacteriastionSpisPinkAbs
Source
Stylophora pistillata. The protein was first extracted and characterized by Alieva et. al. under the name spisCP (GenBank: ABB17971.1). This version is codon optimized for E coli by Genscript.
References
[http://www.ncbi.nlm.nih.gov/pubmed/18648549] Alieva, Naila O., et al. "Diversity and evolution of coral fluorescent proteins." PLoS One 3.7 (2008): e2680.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]