Difference between revisions of "Part:BBa K1028001"
(→Introduction) |
(→Usage and Experimental Data) |
||
| Line 15: | Line 15: | ||
<img src="http://i276.photobucket.com/albums/kk29/JosephBeetBeet/INPSi4Growthcurve.png" style="width:400px;margin-right:160px" ></a> | <img src="http://i276.photobucket.com/albums/kk29/JosephBeetBeet/INPSi4Growthcurve.png" style="width:400px;margin-right:160px" ></a> | ||
| − | <p style="text-align:center"><b>Fig.1:</b> Monitoring the growth rate of cells containing Si4 Binding Peptide attached to the C-terminus of INP, labelled here as Device 2, within the PSB1C3 standard iGEM submission plasmid. Although the graph shows a log and a lag phase for the cells containing the insert plasmid, the absorbance reached in log phase is very low compared to the control cells. | + | <p style="text-align:center"><b>Fig.1:</b> Monitoring the growth rate of cells containing Si4 Binding Peptide attached to the C-terminus of INP, labelled here as Device 2, within the PSB1C3 standard iGEM submission plasmid. Although the graph shows a log and a lag phase for the cells containing the insert plasmid, the absorbance reached in log phase is very low compared to the control cells in fact showing the poorest growth of any of the Leeds 2013 submitted parts. |
</p></html> | </p></html> | ||
Revision as of 12:53, 4 October 2013
INP.Si4 Silica Binding Complex
Introduction
This biobrick was designed utilise Ice Nucleation Protein to express the Si4 Binding Peptide on the extracellular surface of the plasma membrane of E.coli cells.
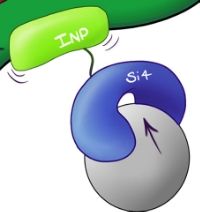
Fig.1 Illustrated schematic of the Si4 Binding Peptide binding presented on the C-terminus of INP as it bind with a silica microsphere.
Usage and Experimental Data
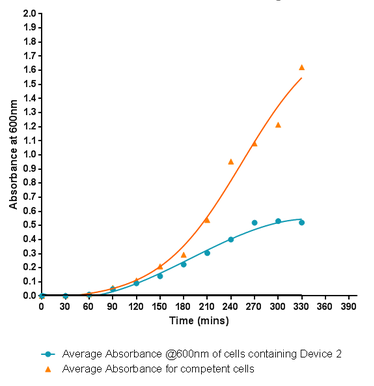
Fig.1: Monitoring the growth rate of cells containing Si4 Binding Peptide attached to the C-terminus of INP, labelled here as Device 2, within the PSB1C3 standard iGEM submission plasmid. Although the graph shows a log and a lag phase for the cells containing the insert plasmid, the absorbance reached in log phase is very low compared to the control cells in fact showing the poorest growth of any of the Leeds 2013 submitted parts.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 409
- 1000COMPATIBLE WITH RFC[1000]
