Difference between revisions of "Part:BBa K1087003"
| Line 4: | Line 4: | ||
This is a strong reporter amilcp with a T7 promoter. When you use T7 polymerase & T7 promoter to control the expression of desired genes,you can ues this reporter since it can be seen by naked eyes in high copy plasmid. Reference BBa_K592025 | This is a strong reporter amilcp with a T7 promoter. When you use T7 polymerase & T7 promoter to control the expression of desired genes,you can ues this reporter since it can be seen by naked eyes in high copy plasmid. Reference BBa_K592025 | ||
| + | |||
| + | ===<h2 style="font-weight: bold;">CSMU_Taiwan 2020 Improve</h2>=== | ||
| + | <html> | ||
| + | Group: CSMU iGEM 2020 | ||
| + | <br> | ||
| + | Author: Huan-Jui Chang, Cheng-Ruei Yang, Hung-Yu Chen <br> | ||
| + | |||
| + | ===Introduction=== | ||
| + | <html> | ||
| + | This year, CSMU wants to solve the problems revolving around diagnosing oral cancer, and we decided to use toehold switches as our detection device. Toehold switches are composed of 3 main parts: trigger binding site, ribosome binding site, and a reporter gene. We wanted to find the most suitable reporter gene which can be measured with ease. Therefore, we had tried different kinds of reporter genes. Including mRFP which has been mentioned in our <a href= | ||
| + | "https://2020.igem.org/Team:CSMU_Taiwan/Contribution">contribution</a>, invertase which is our <a href= "https://2020.igem.org/Team:CSMU_Taiwan/Parts">new part</a>. We also had tried amilCP, which is a blue chromoprotein that is visible to the naked eye. With our improvement, amilCP's expression can be controlled, just like adding an on and off switch. AmilCP would not be expressed without a specific trigger under the regulation of toehold switches. By doing so, it could turn out to be a useful diagnostic tool. | ||
| + | </html> | ||
| + | ===Method=== | ||
| + | <html> | ||
| + | We followed the <a href="https://www.protocols.io/private/170B0BD515DE11EBAE360A58A9FEAC2A">protocol</a> we created. We expressed three of them, the original part ( <a href= "https://parts.igem.org/Part:BBa_K3431049">BBa_K1087003</a>), the toehold switch with the trigger, and the toehold switch without the trigger in vitro environment, using the cell-free PURExpress protein synthesis kits and incubating them for six hours at 37°C. | ||
| + | <br></html> | ||
| + | <br> | ||
| + | <b>Results</b><br> | ||
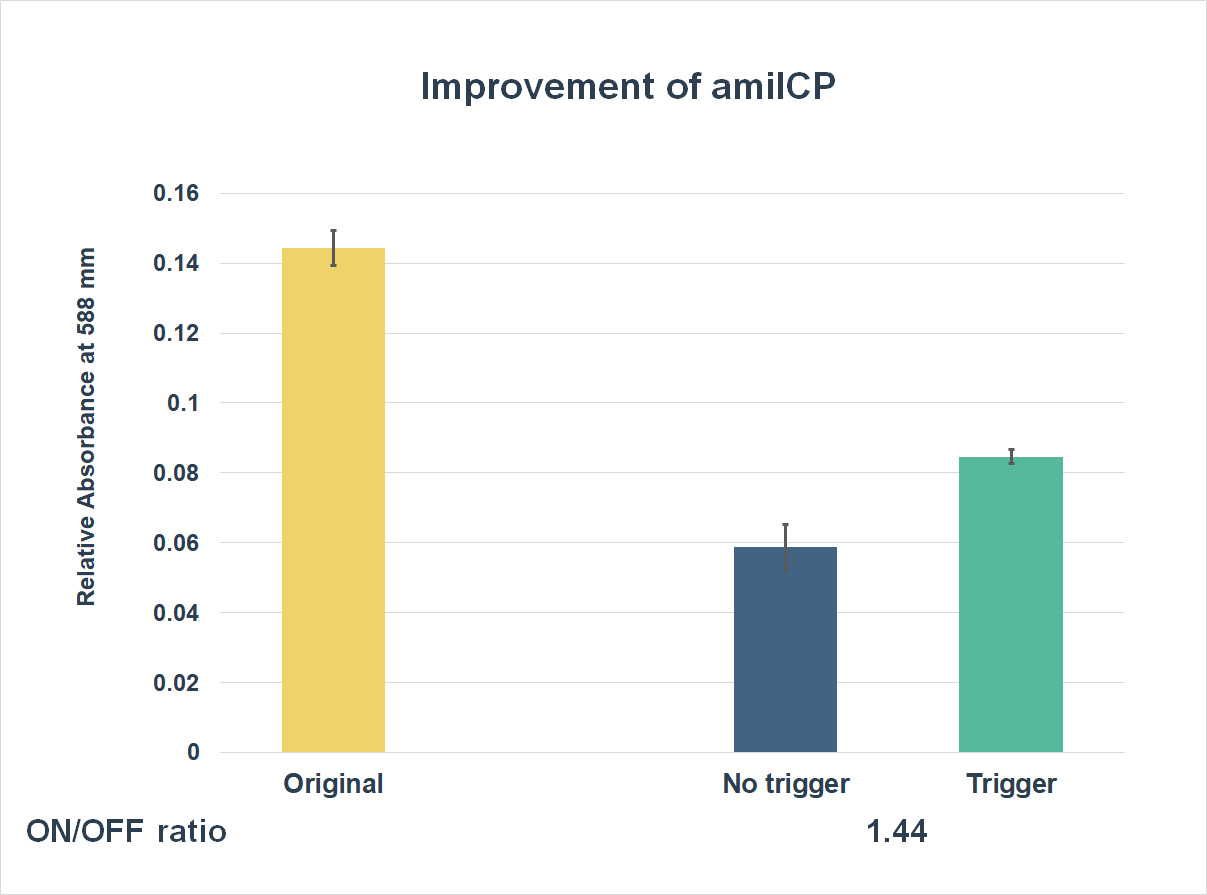
| + | This is the graph that focuses at the sixth hour. It shows the results of our experiments with both parts; one is the original existing part; the other is an improved part. The vertical axis shows the relative absorbing wavelength at 588 mm. And the horizontal axis represents the name of the three. The yellow bar represents the original part, the middle one represents the improved part with a trigger, and the blue one is the improved part without a trigger. | ||
| + | The expression of our improved part is lower than the original ones. However, comparing the middle one and the blue one, the middle one expressed better. The on/ off ratio is 1.44, which were much higher than 1. Representing that there were significant differences between the ON state and the OFF state. The result proved that amilCP with toehold switches could only be triggered by a specific sequence, which also means the amilCP expression is lower in the absence of the trigger. | ||
| + | <html> | ||
| + | <br> | ||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center"> | ||
| + | <img src="https://2020.igem.org/wiki/images/archive/9/9e/20201025012643%21T--CSMU_Taiwan--improve.png" style="width:50%"> | ||
| + | </div> | ||
| + | </html> | ||
| + | <br> | ||
| + | <b>Conclusion</b><br> | ||
| + | We successfully added the new function on amilCP. With our improvement, AmilCP's expression can be controlled, just like adding an on and off switch. AmilCP would not be expressed without a specific trigger. By doing so, it could turn out to be a useful diagnostic tool. | ||
| + | |||
| + | ===Reference Section=== | ||
| + | 1. Alieva, N. O., Konzen, K. A., Field, S. F., Meleshkevitch, E. A., Hunt, M. E., Beltran-Ramirez, V., Miller, D. J., Wiedenmann, J., Salih, A., & Matz, M. V. (2008). Diversity and evolution of coral fluorescent proteins. PloS one, 3(7), e2680. https://doi.org/10.1371/journal.pone.0002680 | ||
| + | |||
| + | 2. Part: BBa_K1087003: https://parts.igem.org/Part:BBa_K1087003 | ||
| + | |||
| + | 3. Part:BBa_K592025: https://parts.igem.org/Part:BBa_K592025 | ||
| + | |||
| + | 4. Pardee, K., Green, A. A., Takahashi, M. K., Braff, D., Lambert, G., Lee, J. W., Ferrante, T., Ma, D., Donghia, N., Fan, M., Daringer, N. M., Bosch, I., Dudley, D. M., O'Connor, D. H., Gehrke, L., & Collins, J. J. (2016). Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell, 165(5), 1255–1266. https://doi.org/10.1016/j.cell.2016.04.059</p> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 09:28, 26 October 2020
T7 promoter-B0034-amilCP-teminator
This is a strong reporter amilcp with a T7 promoter. When you use T7 polymerase & T7 promoter to control the expression of desired genes,you can ues this reporter since it can be seen by naked eyes in high copy plasmid. Reference BBa_K592025
CSMU_Taiwan 2020 Improve
Group: CSMU iGEM 2020
Author: Huan-Jui Chang, Cheng-Ruei Yang, Hung-Yu Chen
===Introduction===
This year, CSMU wants to solve the problems revolving around diagnosing oral cancer, and we decided to use toehold switches as our detection device. Toehold switches are composed of 3 main parts: trigger binding site, ribosome binding site, and a reporter gene. We wanted to find the most suitable reporter gene which can be measured with ease. Therefore, we had tried different kinds of reporter genes. Including mRFP which has been mentioned in our contribution, invertase which is our new part. We also had tried amilCP, which is a blue chromoprotein that is visible to the naked eye. With our improvement, amilCP's expression can be controlled, just like adding an on and off switch. AmilCP would not be expressed without a specific trigger under the regulation of toehold switches. By doing so, it could turn out to be a useful diagnostic tool.
Method
We followed the protocol we created. We expressed three of them, the original part ( BBa_K1087003), the toehold switch with the trigger, and the toehold switch without the trigger in vitro environment, using the cell-free PURExpress protein synthesis kits and incubating them for six hours at 37°C.
Results
This is the graph that focuses at the sixth hour. It shows the results of our experiments with both parts; one is the original existing part; the other is an improved part. The vertical axis shows the relative absorbing wavelength at 588 mm. And the horizontal axis represents the name of the three. The yellow bar represents the original part, the middle one represents the improved part with a trigger, and the blue one is the improved part without a trigger.
The expression of our improved part is lower than the original ones. However, comparing the middle one and the blue one, the middle one expressed better. The on/ off ratio is 1.44, which were much higher than 1. Representing that there were significant differences between the ON state and the OFF state. The result proved that amilCP with toehold switches could only be triggered by a specific sequence, which also means the amilCP expression is lower in the absence of the trigger.
Conclusion
We successfully added the new function on amilCP. With our improvement, AmilCP's expression can be controlled, just like adding an on and off switch. AmilCP would not be expressed without a specific trigger. By doing so, it could turn out to be a useful diagnostic tool.
Reference Section
1. Alieva, N. O., Konzen, K. A., Field, S. F., Meleshkevitch, E. A., Hunt, M. E., Beltran-Ramirez, V., Miller, D. J., Wiedenmann, J., Salih, A., & Matz, M. V. (2008). Diversity and evolution of coral fluorescent proteins. PloS one, 3(7), e2680. https://doi.org/10.1371/journal.pone.0002680
2. Part: BBa_K1087003: https://parts.igem.org/Part:BBa_K1087003
3. Part:BBa_K592025: https://parts.igem.org/Part:BBa_K592025
4. Pardee, K., Green, A. A., Takahashi, M. K., Braff, D., Lambert, G., Lee, J. W., Ferrante, T., Ma, D., Donghia, N., Fan, M., Daringer, N. M., Bosch, I., Dudley, D. M., O'Connor, D. H., Gehrke, L., & Collins, J. J. (2016). Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell, 165(5), 1255–1266. https://doi.org/10.1016/j.cell.2016.04.059</p>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
